Pseudomonas is a genus of aerobic, anaerobic, or facultatively anaerobic, rod-shaped, Gram-negative Gammaproteobacteria in the family Pseudomonadaceae of the phylum Pseudomonadota.
It includes 191 well-characterized species occupying a wide range of ecological niches. They express a heterotrophic nutrition mode and are normally found in soil, water, and vegetation. However, some species are normal microbiota of humans and animals.
Among these nearly 200 known species, 25 are associated with human diseases. Two species; P. aeruginosa and P. maltophilia are associated with more than 80% of the pseudomonads’ human infections. P. aeruginosa is the most frequently reported pathogen of this genus, primarily infecting hospital-admitted patients.
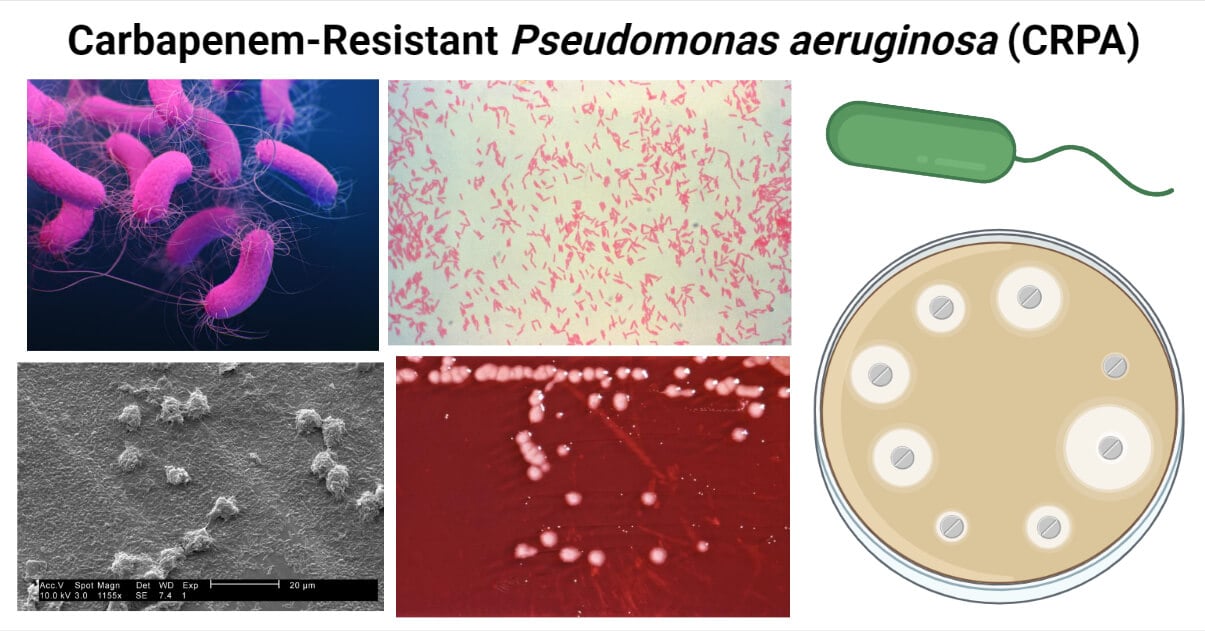
P. aeruginosa is a pathogenic species of this genus. It is responsible mainly for hospital-acquired infections (HAIs).
It is Gram-negative, aerobic or facultative, glucose fermentative, motile, capsulated, and bacilli. Its ability to grow at 42°C and produce green pigmentation (due to pyocyanin pigment) helps identify and differentiate from other Pseudomonas spp.
P. aeruginosa displays a diverse type of clinical manifestation, infecting every tissue in the human body. Although it is an opportunistic pathogen, its infection can be malignant, and associated mortality can be around 50% in the case of P. aeruginosa bacteremia.
Moreover, inherent resistance to diverse antibiotics and rapid development of antibiotic resistance against available options is very high in P. aeruginosa. And this is fueling the case severity and mortality rate associated with P. aeruginosa infections.
It is cosmopolitan in its habitat and is mainly found in soil, water, and decaying vegetation. Due to their capacity to form a capsule and a biofilm, they can survive and colonize in a dry habitat, like medical devices, inanimate surfaces, etc., and infect immune-compromised and hospitalized patients.
Interesting Science Videos
Morphology of Pseudomonas aeruginosa
- Gram-negative Rod-shaped
- 0.5 – 1.0 micron by 1.0 – 3.0 microns (some may grow up to 5.0 microns)
- Single polar flagellum (monotrichous) (some may have 2 – 3 flagella) and contains pili
- Motile
- Non-Capsulated (some strains are capsulated also)
- Non-sporing
Biochemical Characteristics of Pseudomonas aeruginosa
- Facultative anaerobe (mostly aerobic, but can also use nitrate as terminal electron acceptor and thrive in anaerobic conditions)
- Glucose fermentative
- Oxidase positive
- Catalase positive
- IMViC test = -ve, -ve, -ve, +ve (- – – +)
- Urease and DNase negative
- Nitrate positive
- TSI test = Alkaline / Alkaline (Red/Red), no gas, no H2S
- Beta hemolytic
- Pigment-producing (Blue-Green pigmentation)
Cultural Characteristics of Pseudomonas aeruginosa
- Aerobic or facultative incubation with a temperature range from 4-42°C; the optimum temperature of 35 (±2)°C.
- The optimum pH requirement is 6.6 – 7.6, but it can survive in the pH range of 5.6 – 9.0.
- Non-fastidious in nutrient requirement.
Nutrient Agar, MacConkey Agar, and Blood Agar are commonly used in laboratories.
- Nutrient Agar = 2 – 4 mm, Irregular, Convex, Smooth (in fresh culture), Mucoid, Greenish – Bluish, Opaque – Translucent colonies, Sweet fruit-like smell.
- MacConkey Agar = 2 – 3 mm, Circular, Non-lactose fermentative, Low convex – flat, Smooth, Mucoid, Transparent, Colorless colonies, Sweet smell.
- Blood Agar = 2 – 4 mm, Beta-hemolytic, Irregularly circular, Greyish-white, Smooth, Mucoid, Translucent – Opaque colonies, Sweet smell.
- Cetrimide Agar Medium is a selective media; P. aeruginosa produces small 1 – 3 mm, circular, mucoid, smooth, Greenish-blue colonies.
Virulence Factors of Pseudomonas aeruginosa
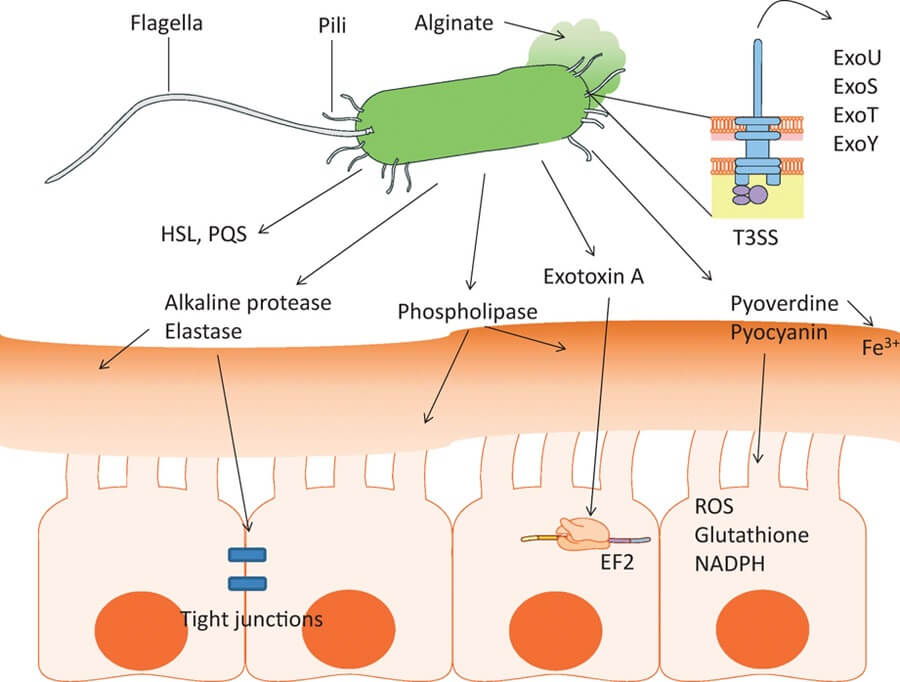
P. aeruginosa contains many cell-associated and extracellular virulence factors contributing to its pathogenicity. The most relevant virulence factors of P. aeruginosa with their proposed roles are summarized in the following table, viz.:
| Virulence Factors | Roles in Pathogenesis |
| Flagella | Helps in biofilm formation Contribute bacterial adhesion and chemotaxis Evoke activation of Toll-like Receptor 5 (TLR5) and immune response |
| Pili (Type IV Pili) | Helps in biofilm formation, adherence, bacterial motility, and exchange of resistance genes Provide resistance against SP-A-mediated phagocytosis |
| Porins They are outer membrane protein channels that regulate bacterial permeability. OprF, OprH, and OprD are the most important porins superfamily conferring virulence to P. aeruginosa. OprF is the major, most abundant, and most important virulence factor. | OprF is responsible for: – Bacterial adhesion with epithelial cells and other bacterial cells – Regulation of other virulence factors – Facilitates biofilm formation and attachment – Provides resistance against antibiotics – Act as a sensor to detect hosts’ immune components – Provides protection against macrophage OprH is responsible for: – Adhesion during respiratory tract infections (RTIs) – Confer resistance against aminoglycosides and polymixins OprD is responsible for: – Adhesions during RTIs – Confer resistance against carbapenems – Aids in biofilm formation OprG and OprQ help in bacterial adhesion during infection OprL protects against oxidative stresses OprJMN and OmpBEG confer antibiotic resistance |
| Lipopolysaccharides (LPS) | LPS are responsible for: – Adherence – Endotoxicity – Induces the production of reactive oxygen species (ROS) and mucin in the respiratory tract – Induce inflammation by stimulating the production of tumor necrosis factor – (TNF – ), Interleukin – 1 and – 6 (IL-1, IL-6), and interferons – Helps in the formation of biofilm and outer membrane vesicles – Confer antibiotic resistance – Inhibit release and action of pore-forming complexes and phagocytes – Protects against oxidative stresses |
| Outer Membrane Vesicles (OMVs) They are outer membrane protein vesicles secreted by the outer membrane of the bacterium for transportation. | OMVs are responsible for: – Bacterial adherence and host recognition – Delivery of virulence factors and sRNAs – Helps in biofilm formation – Helps in evading the host’s immune responses – Acquisition of antibiotic resistance |
| Secreted Toxins, Effector Proteins, and Enzymes P. aeruginosa is found to produce a wide range of toxins and enzymes that are responsible for bacterial pathogenesis. The protein secretory systems are responsible for their production. | Exotoxin A: disrupts protein synthesis in host cells resulting in cell death ExoS and ExoT are effector proteins responsible for: – Apoptosis and necrosis; death of immune cells – Inhibit and disrupt cytoskeleton formation – Inhibit DNA synthesis – Inhibit phagocytosis – Promotes cell adhesions and proliferation ExoU is another effector protein causing the death of phagocytes ExoY is another effector protein disrupting the actin cytoskeleton Proteases are responsible for: – Disrupt immunoglobulins and complements – Degrades fibrinogen and plasminogen; hence, promotes migration in the host body – Inhibit and disrupt antibacterial peptides and phagocytes – Provide resistance against antibiotics Lipases are responsible for: – Aids in laminin binding during RTIs – Show hemolytic activity Pyocyanin is responsible for: – A decline in lung function and severity of infection – Increase H2O2 and ROS content leading to oxidative stress, cell cycle disruption, cell lysis – Degrades enzymes and DNA of infected cells – Reduce ciliary beating increasing mucous secretion and epithelial cell damage in the respiratory tract |
| Protein Secretion System It is molecular machinery that helps form and translocates proteins (toxins and enzymes) and nucleic acids. There are six well-defined types of secretory systems in P. aeruginosa; Type I secretory system (T1SS), Type II secretory system (T2SS), Type III secretory system (T3SS), Type IV secretory system (T4SS), Type V secretory system (T5SS) and Type VI secretory system (T6SS). | T1SS is responsible for the transport of proteases and lipases, adhesions, and colonization; besides, roles in pathogenesis are not known well. T2SS is responsible for biofilm formation, inflammation regulation in hosts, and bacterial motility. Its other roles in pathogenesis are not defined. T3SS plays the following roles: – Quorum sensing Secrets virulence effectors inside the host’s cells disrupt the cell signaling mechanism and cause cell death – Helps escape from the host’s immune responses and colonize, multiply and spread – Confer resistance against Beta-lactam antibiotics T4SS is responsible for: – Horizontal gene transfer of antibiotic resistance genes – Helps in cell adhesion T5SS is responsible for: – Aids in cell motility and biofilm formation T6SS is responsible for: – Promotes bacterial competition and aids in colonization and killing other competing bacteria – Aids in biofilm formation |
| Iron Acquisition System It is the system present in the bacterial cell to meet up their iron requirement to thrive and promote virulence. | – Promotes bacterial survival and proliferation inside the host’s body – Cause inflammation and oxidative damage to host cells |
| Quorum Sensing It is a system of cell-to-cell communication used by the bacterium. P. aeruginosa have four QS systems, via, Las, Rhl, Pqs, and Iqs. | QS is responsible for: – Adaptation inside the host body – Las and Rhl QS systems are involved in the transcription of genes involved in host cell damage and acute infections Induces apoptosis in respiratory epithelial cells – The Pqs system is responsible for the synthesis of hydrogen cyanide, chitinase, and elastase. They also confer antibiotic resistance and release of eDNA – Iqs stimulate apoptosis and inhibit the host’s cell growth – QS system mediates biofilm formation, OMV biogenesis, suppression of immune components, cytotoxicity, etc. |
| Other virulence factors: – Rhamnolipids – Antioxidant Enzymes | – Rhamnolipids degrade lung surfactants, promote biofilm formation and swarming motility, suppress the host’s innate immunity – Antioxidant enzymes protect from oxidative stress while in the host cell. |
Pathogenesis of Pseudomonas aeruginosa
Pseudomonas aeruginosa is an opportunistic pathogen responsible for various acute and chronic human infections, mainly in hospitalized and immune-compromised patients. They can be transmitted via inanimate objects in hospital settings (like medical devices and catheters, fomites, surfaces, etc.), direct contact from person to person, airway transmission through contaminated droplets, and environmental factors like soil, air, food, and water.
Pseudomonas aeruginosa infections are most commonly associated with burns and open wounds, urinary catheters, immunocompromised conditions, elderly, chronic lung diseases like cystic fibrosis, (chronic obstructive pulmonary diseases) COPD and ventilation, diabetes, etc. Pathogenesis also varies according to the site of infection and associated risk factors.
Soon after contacting a suitable host’s surface, the bacteria adhere using their adherence factors like pili, flagella, LPS, OMVs, secretory systems, etc. Once attached to the surface, they express virulence factors to colonize and form a biofilm on that site. The bacterial cells then invade and destroy the host’s cells via cytotoxic effect, provocation of inflammation, apoptosis, and necrosis. They escape the host’s immune components and continue flourishing, invading, and disseminating.
Clinical Manifestation of Pseudomonas aeruginosa
P. aeruginosa is an opportunistic pathogen mainly associated with hospital-acquired infections. Community-acquired infections are also reported but their prevalence is a very low number in comparison to HAIs. They are responsible for acute and chronic infections in individuals with a compromised immune system, COPD, burns and open wounds, diabetes, cystic fibrosis (CF), indwelling catheters and implants, pneumonia, cancer, and traumatic injury.
Biofilm formation and the development of resistance against most of the available antibiotics have made P. aeruginosa a malignant pathogen causing severe and persistent infections with high mortality.
P. aeruginosa is frequently associated with the following medical conditions:
Respiratory Tract Infections
P. aeruginosa is one of the most common bacterial pathogens causing RTIs in hospitalized patients. It causes both acute and chronic RTIs. Pneumonia is the most common clinical syndrome. It is responsible for Ventilator-associated pneumonia (VAP), and pneumonia in patients in an intensive care units (ICUs); mainly in patients with intubation and underlying lung diseases like CF and COPD. Infection remains persistent and results in bilateral, diffused bronchopneumonia with pleural effusion. The mortality rate may be up to 70% in hospitalized CF and COPD patients with diffused bronchopneumonia.
Soft Tissue and Skin Infection
P. aeruginosa is one of the most common bacterial pathogens causing infections in burn and open wounds. Due to the formation of biofilm, pus generation, and resistance against antibiotics, infected wounds don’t heal quickly. Discharge of bluish-greenish pus is the peculiar feature of wound infection by P. aeruginosa.
Besides, they are also associated with chronic paronychia, folliculitis, infected toe web, and cellulitis. Skin infection is seen in those who are frequently exposed to moisture.
Blood Stream Infection (BIs) / Bacteremia
P. aeruginosa is responsible for bacteremia in immunocompromised individuals, mostly hospitalized ones. P. aeruginosa BIs (PA-BSI) are the most serious infections resulting in mortality rates up to 20 – 35% and even more if the pathogen is MDR.
Urinary Tract Infections (UTIs)
Complicated UTIs by P. aeruginosa is often associated with urogenital surgery, indwelling urinary catheters, and kidney stones. The infection needs the removal of such catheters or stones and needs prolonged administration of antibiotics.
Diabetic Foot
P. aeruginosa is often found to cause ulcerated feet in patients with diabetes mellitus, called Diabetic Foot Infection (DFI). DFI is a polymicrobial infection; the dominant bacteria involved are Staphylococcus aureus, Streptococcus spp., and Pseudomonas aeruginosa.
Ear Infections
Otitis externa or ‘Swimmer’s Ear’ is caused by P. aeruginosa, especially in those with water blockage in the ear and any ear injury. Malignant otitis externa is seen in elderly, diabetic, and HIV-AIDS patients. In some cases, the infection may reach up to the temporal bone resulting in skull base osteomyelitis which has a very high mortality rate.
Eye Infections
Pseudomonas aeruginosa Keratitis (corneal inflammation) and endophthalmitis are mainly seen in individuals wearing contact lenses, and underlying conditions like trauma, surgical procedures, or perforation of corneal ulcers.
Central Nervous System Infections
CNS infection by P. aeruginosa is very rare but severe and lethal. It is mainly associated with trauma and surgical procedures.
Infective – endocarditis
P. aeruginosa Endocarditis is rare and often associated with IV drug users or in patients with surgical procedures, IV catheters, and patients who have undergone heart surgery. They usually cause damage to heart valves leading to a heart attack.
What are Carbapenems?
Carbapenems are Broad-spectrum antibiotics used against serious infections, especially those caused by multidrug-resistant bacterial pathogens (MDR Enterobacteriaceae, A. baumannii, Pseudomonas spp., Streptococcus spp., Mycobacterium tuberculosis complex, Haemophilus influenzae, MRSA, Salmonella serovars, etc.)
- β- Lactam antibiotic class capable of inhibiting – Lactamase enzymes
- Can resist the effects of – Lactamase enzymes
- Susceptible to ‘carbapenemases’ and Metallo – β- lactamases
- Have the β- lactam ring with carbon atom replacing sulfur at C-1, and introduction of a double bond between C-2 and C-3 of the ring with the side chains arranged in the trans position.
- The unusual arrangement of side chains in the trans position, unlike in other β- lactam antibiotics having side chains in the cis position, makes it resistant to β- lactamase enzymes such as Extended Spectrum Beta – Lactamases (ESBLs), and AmpC Beta – Lactamases (ABLs).
- Classification: Two groups; a) Group – 1 containing “ERTAPENEM”, b) Group – 2 containing “IMIPENEM”, “MEROPENEM”, and “DORIPENEM”. Others are also available for clinical uses but are not approved and used widely like Panipenem, Biapenem, and Tebipenem.
- Mode of action: Binding to the PBPs (penicillin-binding proteins), hence inhibiting bacterial cell wall formation.
- Called “the antibiotics of last resort”.
What are Carbapenem-Resistant Organisms (CROs)?
- Bacteria that have developed several mechanisms to protect themselves from the inhibitory or bactericidal action of carbapenems are carbapenems-resistant organisms.
- CROs are emerging and disseminating globally and has risen as a pathogen causing some of the most serious health problem and the most dangerous threat to public health.
- They are labeled as organisms of “urgent threat requiring urgent countermeasures” by the US Center for Disease Control and Prevention (CDC), and “priority one pathogen” by the World Health Organization (WHO).
Some most serious carbapenem-resistant bacteria are:
- Carbapenem-resistant Enterobacteriaceae (CRE)
[Carbapenem-resistant Klebsiella pneumoniae is the most important among them] - Carbapenem-resistant Acinetobacter baumannii (CRAB)
- Carbapenem-resistant Pseudomonas aeruginosa (CRPA)
What is Carbapenem-resistant Pseudomonas aeruginosa?
- CRPA is a group of P. aeruginosa bacteria that have developed resistance against carbapenem antibiotics. CRPA is labeled as “a serious threat” by the US CDC in 2019 and “a priority 1 – critical pathogen” by the WHO since 2017. It is also a member of the ESKAPE group including MDR pathogenic bacteria associated with most of the HAIs.
- The limited availability of antibiotics to treat has made CRPA a very serious pathogen. Infections by CRPA are difficult to treat, need longer antibiotic courses and hospital stays, and have higher morbidity and mortality.
- P. aeruginosa were not naturally resistant to carbapenems. In due course of time, due to repeated exposure to the carbapenems and the acquisition of resistant genes from other bacteria, P. aeruginosa has genetically evolved to resist the inhibitory and bactericidal effects of carbapenems. They have developed several mechanisms like structural modification, development of virulence factors, and production of enzymes to resist the carbapenems.
Risk Factors for CRPA Infections
- Hospitalized patients
- Immunocompromised patients
- Patients with indwelling catheters (indwelling urinary catheters)
- Patients with intubation and ventilation
- Patients with invasive medical devices like IV catheters
- Burn and open wound patients
- Diabetic, cancer, and AIDS patients
- Patients with underlying lung diseases like CF and COPD.
Mechanisms of Resistance Against Carbapenems by CRPA
P. aeruginosa has multiple mechanisms of resistance against carbapenems. Some of the well-studied resistant mechanisms are:
Enzymatic Modification of Antibiotics
CRPA can produce different carbapenemase enzymes that can hydrolyze the β- lactam ring of carbapenems, making them ineffective.
P. aeruginosa is found to produce all types of transferable carbapenemases except the Seoul Imipenemase (SIM – 1) enzyme.
- The “Metallo – – Lactamases (MBLs)” is the most important carbapenemases produced by many of the clinical isolates. Imipenem-resistant Pseudomonas (IMP) Metallo-β-lactamase group and Verona integron-related Metallo-β-lactamase (VIM) group are the most frequently reported MBLs types.
- The “K. pneumoniae carbapenemase (KPC) β- lactamases” are also detected in the P. aeruginosa isolates. However, they are mainly reported from the Latin American region, and sporadically from other continents.
- The class – D β- lactamases like “Oxacillinase (OXA β- lactamases)” are rarely found in P. aeruginosa.
Modification in Membrane Permeability
Entry of carbapenems inside the cell of P. aeruginosa is primarily through the OprD outer membrane porin channel. Reduced expression of the OprD porin channel is seen in CRPA. The downregulation of the OprD porin is linked with a mutation in the OprD genes of the CRPA. This reduced expression or loss of the OprD porin channel confers resistance against the “IMIPENEM” and “MEROPENEM”.
Over Expression of Efflux Pumps
Over-expression of the efflux pumps is the most important strategy adopted by P. aeruginosa to confer resistance against the carbapenems.
Multidrug efflux system AB – outer membrane protein M (MexAB – OprM) is the most important efflux pump responsible for carbapenems resistance in CRPA.
Multidrug efflux system XY – outer membrane protein M (MexXY – OprM) is another important one, which together with the MexAB – OprM confer MDR to P. aeruginosa. MexCD – OprJ, MexEF-OprN, MexJK-OprM, and MexVW-OprM are other efflux systems that are found to be responsible for resistance against “MEROPENEM” and other antibiotic classes, but are not able to expel “IMIPENEM”.
Horizontal Gene Transfer
HGT is an adaptive mechanism where the genes conferring resistance to carbapenems are transferred from the environment or other carbapenems-resistant bacteria to susceptible P. aeruginosa.
Biofilm Formation
The formation of biofilm is an important adaptive mechanism that provides resistance against a wide range of antibiotics including carbapenems. Biofilm act as a physical barrier that protects antibiotics from entering the cell cytoplasm. Also, they help in forming persistent cells that can cope with the antibiotics.
Epidemiology of Carbapenem-resistant Pseudomonas aeruginosa
CRPA is reported globally, especially found to be associated with HAIs. In the USA ~10 – 30% of the clinical isolates of P. aeruginosa are found to be resistant to one or more carbapenems. A report from the American Society of Microbiology (ASM) showed that P. aeruginosa and S. aureus are the most common MDR pathogen distributed globally. The prevalence of CRPA is higher in the Asia-Pacific region, Europe, and Latin America. The report also emphasizes that the susceptibility of Meropenem against P. aeruginosa is decreasing rapidly.
Identification of Carbapenem-resistant Pseudomonas aeruginosa
For identification of CRPA, first, the isolates are identified as P. aeruginosa using biochemical tests or molecular methods (PCR, gene sequencing, DNA Probe), then, the P. aeruginosa are studied for their susceptibility against the carbapenems using phenotypic or molecular approaches.
A biochemical testing algorithm is the most commonly used method in a common laboratory for the identification of P. aeruginosa.
Biochemical Algorithm For Identification of P. aeruginosa
- Aerobic (facultative anaerobe), Non-fastidious, Optimum growth at 370C
- Gram-negative Rod
- Motile
- Catalase and Oxidase +ve
- IMViC = – – – +
- Nitrate +ve
- Urease non-reducing (- ve)
- TSI = R/R (Alkaline / Alkaline), Gas – ve and H2S –ve
- Cetrimide test +ve
- Pigmentation +ve (bluish-green pigmentation)
- Fruit (grape) like odor (due to production of aminoacetophenone)
- Beta hemolytic
- Acylamidase test +ve
Confirmation of Carbapenem Resistance in Pseudomonas aeruginosa (CRPA)
Phenotypic Confirmation of CRPA
Phenotypically, CRPA can be confirmed by performing Antimicrobial Sensitivity Test using carbapenems. If the size of the zone of inhibition is less than recommended or in the level of resistance, then we can confirm the isolate to be CRPA.The zone size interpretative chart for carbapenems against P. aeruginosa according to the CLSI standard is presented below.
| Carbapenem Antibiotics | Zone Size (in mm) For: | Zone Size (in mm) For: | Zone Size (in mm) For: |
| Sensitive | Intermediate | Resistance | |
| Doripenem (DOR 10 mcg) | ≥19 | 16 – 18 | ≤15 |
| Ertapenem (ETP 10 mcg) | Not available | Not available | Not available |
| Imipenem (IMP 10 mcg) | ≥19 | 16 – 18 | ≤15 |
| Meropenem (MRP 10 mcg) | ≥19 | 16 – 18 | ≤15 |
The zone size interpretative chart for carbapenems against P. aeruginosa according to the EUCAST standard is presented below.
| Carbapenem Antibiotics | Zone Size (in mm) For: | Zone Size (in mm) For: | Zone Size (in mm) For: |
| Sensitive | Intermediate | Resistance | |
| Doripenem (DOR 10 mcg) | ≥25 | 22 – 24 | ≤22 |
| Ertapenem (ETP 10 mcg) | Not available | Not available | Not available |
| Imipenem (IMP 10 mcg) | ≥20 | 17 – 19 | ≤17 |
| Meropenem (MRP 10 mcg) | ≥24 | 18 – 23 | ≤18 |
The common methods for phenotypic detection of carbapenemase production in CRPA include:
- Modified Hodge Test
- Carba NP Test
- Modified Carbapenem Inactivation Method
- Blue Carba Test
- mCIM/eCIM Test
- Double Disk Synergy Test (using 10 μg 0.5 M EDTA disk and 10 μg imipenem or meropenem disk) and Combined Disk Synergy Test (using one imipenem or meropenem disk with EDTA and another without EDTA)
Molecular Confirmation of CRAB
- PCR is the most widely used tool in laboratories to identify (- lactamase genes) genes producing carbapenemases and modified porins and efflux pumps.
Xpert Carba-R molecular test is a common PCR method for the detection of carbapenemase production.
Multiplex PCR for the presence of blaKPC, blaNDM-1, blaIMP, blaVIM, blaSIM, blaSPM, blaGIM, and blaOXA genes are routinely used. - Duplex multiple cross displacement amplification combined with lateral flow biosensor (MCDA-LFB) method is a new and reliable method.
- DNA Microarray is another commonly used method.
Prospective Treatment Options for CRPA Infections
Carbapenems are “the antibiotic of last resort”, so treating an infection with CRPA is a very difficult task. There is a limited treatment option for CRPA infection. The Infectious Disease Society of America (IDSA) has released guidelines for the treatment of Difficult to Treat Resistant (DTR) Pseudomonas aeruginosa (DTR P. aeruginosa is defined as P. aeruginosa that are resistant to piperacillin-tazobactam, ceftazidime, cefepime, aztreonam, meropenem, imipenem-cilastatin, ciprofloxacin, and levofloxacin) including CRPA and it includes the following treatment options: (reference: doi: 10.1093/cid/ciac268. PMID: 35439291)
- Polymyxins (Colistin and Polymyxin B)
- Ceftolozane-tazobactam,
- Ceftazidime-avibactam,
- Imipenem-cilastatin-relebactam,
- Cefiderocol,
- Meropenem – Vaborbactam
- A single dose of an aminoglycoside
- Ampicillin – Sublactam (Cefoperazone – Sulbactam)
- Plazomicin
Future/Alternative Treatment Options Under Study
- Cefederocol and Fosfomycin
- Delafloxacin (Baxdale)
- Nanoparticle therapy
- Phage therapy (phage cocktail)
- Nucleic acid-based antibiotics (like plasmid DNA, single-stranded and double-stranded DNA (ssDNA and dsDNA), and ssRNA)
- Engineered endolysins
- Vaccines (Protein vaccines and mRNA/DNA vaccines)
Prevention and Control Measures from CRPA Infections
- Multimodal infection prevention and control strategy in community. This includes methods to prevent being infected by P. aeruginosa; such as Hygiene maintenance, sanitation, contact precaution, and patient isolation.
- Rigorous hand hygiene practice in hospital settings
- Proper maintenance of sanitation in patient rooms and proper sterilization of hospital settings
- Use of proper PPE by hospital staff and regular sanitation practice before handling a new patient
- Sterilization of medical devices and ensuring their safety before administration
- Effective surveillance system
- Maintaining a sterile environment in pre- and post-operation wards, operation theater, ICUs, and ventilators
- No antibiotics without prescription
- Rational prescription and use of antibiotics
- Antimicrobial therapy stewardship
References
- Jurado-Martín, I., Sainz-Mejías, M., & McClean, S. (2021). Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. International Journal of Molecular Sciences, 22(6). https://doi.org/10.3390/ijms22063128
- Iglewski BH. Pseudomonas. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 27. Available from: https://www.ncbi.nlm.nih.gov/books/NBK8326/
- Stratton CW. Pseudomonas aeruginosa. Infect Control. 1983 Jan-Feb;4(1):36-40. doi: 10.1017/s0195941700057647. PMID: 6403483.
- Diggle, S. P., & Whiteley, M. (2020). Microbe Profile: Pseudomonas aeruginosa: opportunistic pathogen and lab rat. Microbiology, 166(1), 30-33. https://doi.org/10.1099/mic.0.000860
- Morphology & Culture Characteristics of Pseudomonas aeruginosa (paramedicsworld.com)
- Pseudomonas aeruginosa -Gram Stain, Culture Characteristics, Infection (microscopemaster.com)
- Mielko, K. A., Jabłoński, S. J., Milczewska, J., Sands, D., Łukaszewicz, M., & Młynarz, P. (2019). Metabolomic studies of Pseudomonas aeruginosa. World Journal of Microbiology & Biotechnology, 35(11). https://doi.org/10.1007/s11274-019-2739-1
- Al-Wrafy, Fairoz & Brzozowska, Ewa & Górska, Sabina & Gamian, Andrzej. (2017). Pathogenic factors of Pseudomonas aeruginosa – the role of biofilm in pathogenicity and as a target for phage therapy. Postępy Higieny i Medycyny Doświadczalnej (Advances in Hygiene and Experimental Medicine). 71. 78-91. 10.5604/01.3001.0010.3792.
- Vivo, A., Fitzpatrick, M.A., Suda, K.J. et al. Epidemiology and outcomes associated with carbapenem-resistant Acinetobacter baumannii and carbapenem-resistant Pseudomonas aeruginosa: a retrospective cohort study. BMC Infect Dis 22, 491 (2022). https://doi.org/10.1186/s12879-022-07436-w
- Shaan L. Gellatly, Robert E.W. Hancock, Pseudomonas aeruginosa : new insights into pathogenesis and host defenses , Pathogens and Disease, Volume 67, Issue 3, April 2013, Pages 159–173, https://doi.org/10.1111/2049-632X.12033
- Andreu Coello Pelegrin;Mattia Palmieri;Caroline Mirande;Antonio Oliver;Pieter Moons;Herman Goossens;Alex van Belkum; (2021). Pseudomonas aeruginosa: a clinical and genomics update . FEMS Microbiology Reviews, (), –. doi:10.1093/femsre/fuab026
- Mariani-Kurkdjian P, Bingen E. Pseudomonas aeruginosa: résistances aux antibiotiques [Pseudomonas aeruginosa: resistance to antibiotics]. Arch Pediatr. 2006 Oct;13 Suppl 1:S5-9. French. PMID: 17370389.
- Liao, C., Huang, X., Wang, Q., Yao, D., & Lu, W. (2022). Virulence Factors of Pseudomonas Aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Frontiers in Cellular and Infection Microbiology, 12. https://doi.org/10.3389/fcimb.2022.926758
- Tenover, F. C., Nicolau, D. P., & Gill, C. M. (2022). Carbapenemase-producing Pseudomonas aeruginosa –an emerging challenge. Emerging Microbes & Infections, 11(1), 811-814. https://doi.org/10.1080/22221751.2022.2048972
- Tenover, Fred & Gill, Christian & Nicolau, David. (2022). Carbapenemase-Producing Pseudomonas aeruginosa –an Emerging Challenge: Carbapenemases in Pseudomonas aeruginosa. Emerging Microbes & Infections. 11. 1-9. 10.1080/22221751.2022.2048972.
- Pai, H., Kim, W., Kim, J., Lee, J. H., Choe, K. W., & Gotoh, N. (2001). Carbapenem Resistance Mechanisms in Pseudomonas aeruginosa Clinical Isolates. Antimicrobial Agents and Chemotherapy, 45(2), 480-484. https://doi.org/10.1128/AAC.45.2.480-484.2001
- Meletis, G., Exindari, M., Vavatsi, N., Sofianou, D., & Diza, E. (2012). Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia, 16(4), 303-307. https://doi.org/https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3738602/
- Arai, T., Otake, M., Enomoto, S., Goto, S. and Kuwahara, S. (1970), Determination of Pseudomonas aeruginosa by Biochemical Test Methods. Japanese Journal of Microbiology, 14: 279-284. https://doi.org/10.1111/j.1348-0421.1970.tb00525.x
- Biochemical Test and Identification of Pseudomonas aeruginosa (microbiologyinfo.com)
- Biochemical Test of Pseudomonas aeruginosa (microbenotes.com)
- de Oliveira Santos IC, da Conceiçāo Neto OC, da Costa BS, Teixeira CBT, da Silva Pontes L, Silveira MC, Rocha-de-Souza CM, Carvalho-Assef APD. Evaluation of phenotypic detection of carbapenemase-producing Pseudomonas spp. from clinical isolates. Braz J Microbiol. 2022 Nov 3. doi: 10.1007/s42770-022-00857-4. Epub ahead of print. PMID: 36327041.
- Verma, N., Prahraj, A. K., Mishra, B., Behera, B., & Gupta, K. (2019). Detection of carbapenemase-producing Pseudomonas aeruginosa by phenotypic and genotypic methods in a tertiary care hospital of East India. Journal of Laboratory Physicians, 11(4), 287-291. https://doi.org/10.4103/JLP.JLP_136_19
- Mohanty S, Maurya V, Gaind R, Deb M. Phenotypic characterization and colistin susceptibilities of carbapenem-resistant of Pseudomonas aeruginosa and Acinetobacter spp. J Infect Dev Ctries. 2013 Nov 15;7(11):880-7. doi: 10.3855/jidc.2924. PMID: 24240048.
- Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious Diseases Society of America 2022 Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin Infect Dis. 2022 Aug 25;75(2):187-212. doi: 10.1093/cid/ciac268. PMID: 35439291.
- Doi, Y. (2019). Treatment Options for Carbapenem-resistant Gram-negative Bacterial Infections. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America, 69(Suppl 7), S565. https://doi.org/10.1093/cid/ciz830
- Tracking Carbapenem-Resistant Pseudomonas aeruginosa | HAI | CDC
- Walters, M., Grass, J. E., Bulens, S. N., Hancock, E. B., Phipps, E. C., Muleta, D….Kallen, A. (2019). Carbapenem-Resistant Pseudomonas aeruginosa at US Emerging Infections Program Sites, 2015. Emerging Infectious Diseases, 25(7), 1281-1288. https://doi.org/10.3201/eid2507.181200.
- Rossi Gonçalves I, Dantas RCC, Ferreira ML, Batistão DWDF, Gontijo-Filho PP, Ribas RM. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2017 Apr-Jun;48(2):211-217. doi: 10.1016/j.bjm.2016.11.004. Epub 2016 Nov 26. PMID: 28034598; PMCID: PMC5470431.
- Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in health care facilities (who.int)

thank you valuable information