Interesting Science Videos
Deoxyribonuclease (DNase) Test Definition
Deoxyribonuclease (DNase) Test is a biochemical test performed to differentiate organisms on the basis of their ability to produce the DNase enzyme.
- Deoxyribonucleic acid (DNA) is a large molecular-sized polymer composed of multiple nucleotides monomer that is large in size and thus, cannot enter the bacterial cell membrane.
- Microorganisms produce the deoxyribonuclease enzyme to breakdown the DNA into smaller monomers which can then be taken into the cell easily.
- The nucleotides are used to make nucleic acid for the bacteria as well as a source of nitrogen, phosphorus, and carbon.
- Some microorganisms can even produce extracellular DNase that breaks down larger DNA into smaller monomeric units so that they can be taken into the cell via the transport proteins present on the cell membrane.
- The degradation of DNA is also considered a virulence factor as it causes the degradation of the host’s DNA.
- The ability to produce DNase can thus be used to differentiate between different pathogenic microorganisms.
- DNase test is an important test for the presumptive identification of Staphylococcus aureus and differentiates it from other Staphylococcal species.
Objectives of DNase Test
- To determine the ability of an organism to produce the DNase enzyme.
- To differentiate and identify S. aureus from other Staphylococcal species.
Principle of DNase Test
- DNases are enzymes that hydrolyze DNA and release free nucleotides and phosphate.
- The deoxyribonuclease enzyme produced by bacteria is extracellular endonucleases that break down DNA, yielding a high concentration of oligonucleotides.
- The media used to detect these enzymes can be made by using various indicators (toluidine blue or methyl green) or no indicators to detect the hydrolysis of DNA.
- The first method is performed with no indicator. The hydrolysis of DNA is indicated by the clearing of the agar after the addition of HCl (the oligonucleotides dissolve in acid causing a clear zone, but DNA salts are insoluble).
- When methyl green indicator is added, DNA combines with the methyl green to produce a green color.
- The complex is released when the DNA is hydrolyzed, and the freed methyl green is colorless at pH 7.5.
- When toluidine blue O (TBO) is added, a complex is formed with the DNA, which changes its structure when DNA is hydrolyzed, resulting in a bright pink color.
- The media with dyes can inhibit some microbial growth. Using a heavy inoculum prevents this problem and makes the test more rapid, as it detects preformed enzymes.
- Staphylococcus aureus possesses a heat-stable enzyme, a thermonuclease. To detect this enzyme, first, the organisms are destroyed by heat, and then the free DNase reacts with the medium.
Microorganisms Tested
- Gram-negative bacilli that are suggestive of Stenotrophomonas maltophilia (positive) and are colistin or polymyxin B resistant to separate from Burkholderia cepacia (negative).
- Gram-negative diplococci that are presumptive for Moraxella catarrhalis (positive).
- Gram-positive cocci that are presumptive for S. aureus (positive) and are difficult to separate from other closely related Staphylococcal species and have a questionable coagulase reaction. Some staphylococci might not grow on media with dyes, so the method without the indicator may be used.
- Enterobacteriaceae to identify Serratia (positive) and separate them from Klebsiella spp. and Enterobacter spp. Serratia fonticola is the only Serratia sp. that is negative for DNase.
- Oxidase-positive, indole-positive, Gram-negative rods in order to separate Aeromonas and Vibrio cholerae (DNase positive) from Plesiomonas shigelloides (DNase negative).
Media, Reagents, and Supplies Used
Media Used
- DNase Test Agar Base is used for testing the production of the DNase enzyme. The medium can also be added with indicators like methyl green or toluidine blue O.
- The composition of the DNase Test Agar Base is given below:
| S.N | Ingredients | Gram/liter |
| 1. | Tryptone | 15.0 |
| 2. | Soya peptone | 5.0 |
| 3. | Deoxyribonucleic acid (DNA) | 2.0 |
| 4. | Sodium chloride | 5.0 |
| 5. | Bacteriological agar | 15.0 |
| Final pH at 25°C: 7.3 ±0.2 |
Reagent Used
- 1 N HCl
Supplies Used
- Sterile sticks, needles, or inoculating loops
- Pasteur pipettes or drinking straws
- Boiling heat block
- Incubators at 35 and 30°C
Procedure of DNase Test
A. Preparation of the media
- In a beaker, 42 grams of the dehydrated powder or lab-prepared media is added to 1000 milliliters of pure distilled or deionized water.
- The suspension is heated with agitation to bring it to boil in order to dissolve the medium completely.
- The dissolved medium is then autoclaved at 15 lbs pressure (121°C) for 15 minutes.
- Once the autoclaving process is complete, the beaker is taken out and cooled to a temperature of about 40-45°C.
- The media is then poured into sterile Petri plates under sterile conditions.
- If indicators are to be added, 0.1 gram of toluidine O or methyl green is added to the medium before sterilizing.
B. DNase Test
- Without indicator
- The agar plates are inoculated with the test organism from an 18-hour culture with a sterile inoculating loop or needle.
- The plates are incubated at 35-37°C for 24 hours.
- The incubated agar plates are flooded with a 1N HCl solution, and the excess acid is tipped off.
- Some time is allowed for the reagent to be absorbed into the plates.
- The plates are observed for a clear zone around the colonies within 5 minutes.
- With indicator
- The agar plates with an indicator are inoculated with the test organism from an 18-hour culture with a sterile inoculating loop or needle.
- The plates are incubated for 24 hours at 35-37°C.
- The plates are then observed for the change in color of the indicator.
Result Interpretation of DNase Test
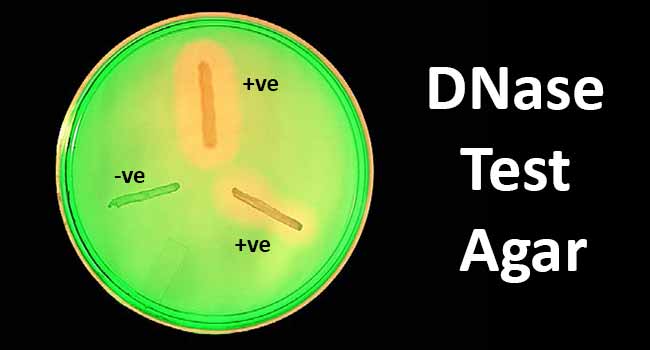
Figure: DNA hydrolysis. Positive- Staphylococcus aureus; Positive- Serratia marcescens; and Negative. Image Source: Bailey and Scott’s Diagnostic Microbiology. Elsevier.
Without indicator
- A clear zone around the colonies demonstrates a positive test after the addition of 1N HCl.
- A negative test is the absence of a clear zone after the addition of 1N HCl.
With indicator (Toluidine Blue O)
- A positive test is demonstrated by the development of a pink or red halo around the colony or the well in the agar.
- A negative test is demonstrated by no change in the royal blue color of the medium.
| S.N | Organism | Growth | Result |
| 1. | Serratia marcescens | Luxuriant | Positive test; change in color from blue to pink around the colonies when toluidine blue is used / clear zone around the colonies when plates are flooded with 1N HCl. |
| 2. | Staphylococcus aureus subsp. aureus | Luxuriant | Positive test; change in color from blue to pink around the colonies when toluidine blue is used / clear zone around the colonies when plates are flooded with 1N HCl. |
| 3. | Staphylococcus epidermidis | Good | Negative; no change in color/ np clearing around the colonies. |
| 4. | Streptococcus pyogenes | Luxuriant | Positive test; change in color from blue to pink around the colonies when toluidine blue is used / clear zone around the colonies when plates are flooded with 1N HCl. |
Uses of DNase Test
- DNase test is used to detect the ability of an organism to produce the DNase enzyme.
- This test can also be used to differentiate S. aureus from other Staphylococcal species.
- It is used to differentiate Serratia spp. as it produces a DNase, which separates non-pigmented strains from most other Enterobacteriaceae.
Limitations of DNase Test
- Broad or large inoculum may result in complete decolorization of the media, as a result of the reduction of the dye. If this case, the test must be repeated.
- Medium with methyl green is better for organisms, such as Gram-negative rods, that first grow on the medium and then demonstrate a positive test.
- For Moraxella and Gram-positive cocci with Toulidine green O testing, a low inoculum can result in a false-negative test, since these organisms may not grow well on the medium.
References and Sources
- Biochemical Tests for the Identification of Aerobic Bacteria. (2016). Clinical Microbiology Procedures Handbook, 3.17.1.1–3.17.48.3.DOI:10.1128/9781555818814.ch3.17.1
- DNase Test Agar Base. M482. HiMedia Laboratories.
- 2% – https://microbenotes.com/citrate-utilization-test-principle-procedure-and-result-interpretation/
- 11% – http://universe84a.com/dnase-test/
- 1% – https://microbeonline.com/deoxyribonuclease-dnase-test-principle-procedure-results/
- 1% – https://himedialabs.com/TD/M482.pdf
- <1% – https://www.reference.com/science/agar-plates-turned-upside-down-incubated-dea80e0278095dbf
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC271612/
- <1% – https://quizlet.com/77236037/microbiology-lab-dna-hydrolysis-test-flash-cards/
- <1% – https://quizlet.com/127518332/micro-lab-quiz-ex-1214-flash-cards/
- <1% – https://microbenotes.com/coagulase-test-principle-procedure-and-result-interpretation/
- <1% – https://en.wikipedia.org/wiki/Agar_plate
- <1% – https://edusanjalbiochemist.blogspot.com/2013/01/urinalysis-chemical-examination.html
