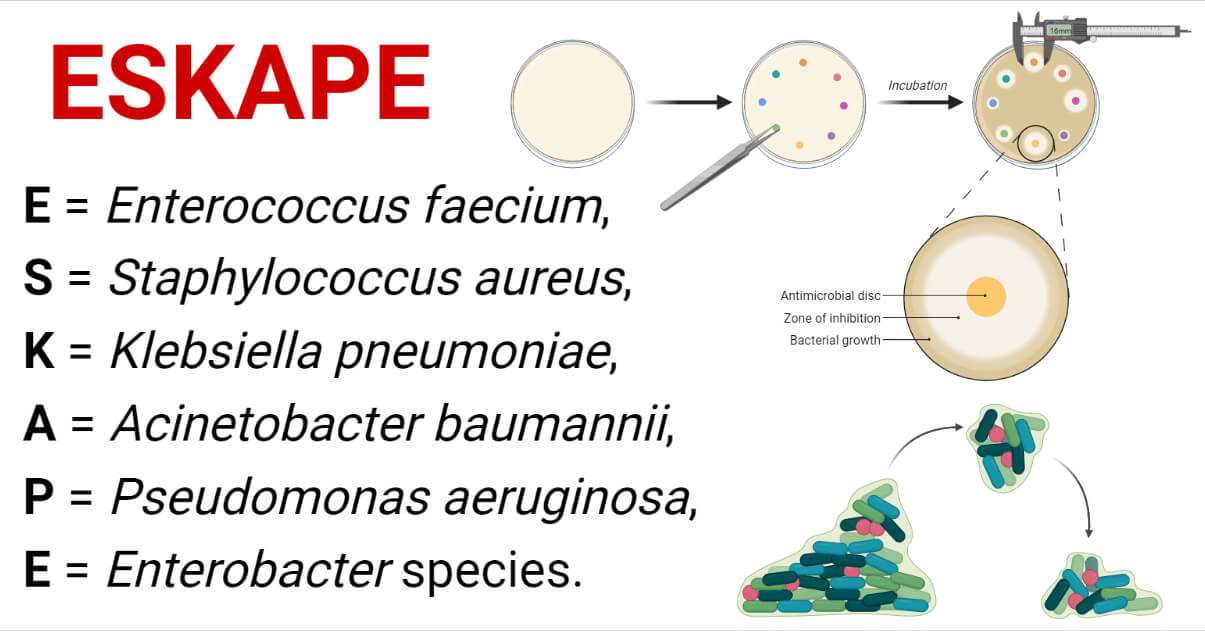
ESKAPE Pathogens are a group of multi-drug resistant pathogenic bacteria, mostly responsible for nosocomial (hospital-acquired) infections. ESKAPE is a group of 6 highly pathogenic bacteria associated with severe nosocomial infections.
ESKAPE is an acronym that stands for :
E = Enterococcus faecium,
S = Staphylococcus aureus,
K = Klebsiella pneumoniae,
A = Acinetobacter baumannii,
P = Pseudomonas aeruginosa, and
E = Enterobacter species.
Antimicrobial-resistant pathogens have been described as serious threats to human health since the early 2000s. The emergence of drug-resistant pathogens has increased the severity and frequency of infections, thus demanding urgent focus on developing new effective antimicrobials. To systematize the surveillance and research on these emerging drug-resistant pathogens, the World Health Organization (WHO) published a list of pathogens for which new effective treatment was urgently needed in February 2017. This broad list contains a group of pathogenic bacteria under “priority status” and named “ESKAPE”.
Before the WHO, the Infectious Disease Society of America (IDSA) had identified these bacteria as major causes of nosocomial infection in 2004. But, ESPAKE became a topic of concern only drug resistance was taken seriously, and the WHO has alerted about the threat of antimicrobial resistance.
Most of the bacteria in this group are listed by the CDC (Center for Disease Control and Prevention) in the CDC’s 2019 Antimicrobial Resistant Threat List. Acinetobacter and Enterobacterales (Klebsiella, Enterobacter) are listed under the urgent threat list, while Pseudomonas, Staphylococcus, and Enterococci are listed under the serious threat list.
Interesting Science Videos
List of ESKAPE Pathogens
ESKAPE contains multi-drug resistant strains of the following six pathogenic bacteria:
1. Enterococcus faecium
- It is a facultatively anaerobic, lactic acid fermenting, non-hemolytic, gram-positive cocci bacteria in the genus Enterococcus of the family Enterococcaceae of the phylum Bacillota.
- Being a part of the human microbiome, it is found in the gastrointestinal tract (GI tract) of humans. However, they have developed multi-drug resistance and are now frequently reported to cause infections in hospitalized patients. They can develop biofilm and hence are involved in medical device-associated infections.
- The special focus is on Vancomycin-resistant strains, called the Vancomycin-Resistant E. faecium (VR E. faecium).
- Vancomycin-resistant strains of E. faecium are reported as a major cause of device-associated infections like ventilator-associated pneumonia and other respiratory tract infections (RTIs), catheter-associated UTIs, catheter-associated surgical wound infections, and bloodstream infections.
2. Staphylococcus aureus
- S. aureus is a gram-positive, catalase-positive, facultatively anaerobic, cocci bacterium in the genus Staphylococcus of the family Staphylococcaceae of the phylum Bacillota.
- It is the most abundant normal flora of the skin and nasal cavity. However, it is frequently reported as opportunistic pathogens causing skin infections, food poisoning, UTIs, bacteremia, sepsis, RTIs, etc.
- S. aureus can develop biofilm in medical devices and are the most common bacteria in the body of every people. Hence they account for the majority of nosocomial infections worldwide. Due to resistance to most available treatments, S. aureus is now emerging as a serious threat in hospital settings, causing an increase in mortality in the cases of S. aureus infection.
- Methicillin-Resistant Staphylococcus aureus (MRSA) is the most common drug-resistant strain of S. aureus, accounting for almost 50% of Staphylococcal infections. MRSA is regarded as a superbug and is frequently associated with skin and soft tissue infections, UTIs, and sepsis.
3. Klebsiella pneumoniae
- K. pneumoniae is a non-motile, oxidase-negative, gram-negative rod-shaped bacterium of the Enterobacteriaceae family.
- It is found in the human body in low numbers as normal flora of the GI tract and skin. Despite being a commensal, it is one of the most common pathogens causing bacterial pneumonia. It is mainly responsible for pneumonia in patients in ventilators and ICUs, but it can also cause UTIs, surgical wound infections, and catheter-associated infections.
- Carbapenem-Resistant K. pneumoniae (CRKP) is the pathogen on the urgent threat list.
4. Acinetobacter baumannii
- A. baumannii is a species of aerobic, glucose non-fermentative, Cocco-bacilli, Gram-negative Gammaproteobacteria in the genus Acinetobacter of the family Moraxellaceae of phylum Pseudomonadota.
- It is normally found in soil and water and as transient flora in human skin. Nosocomial infections by A. baumannii, specially RTIs, UTIs, and wound infections, are increasing rapidly.
- One of the most dangerous strains of A. baumannii is Carbapenem-Resistant A. baumannii (CRAB) which is under the urgent threat list of WHO and CDC. CRAB is mainly associated with ventilator-associated pneumonia (VAP), UTIs, and wound infection in hospitalized patients.
5. Pseudomonas aeruginosa
- P. aeruginosa is a Gram-negative, rod-shaped, encapsulated, facultative anaerobic Gammaproteobacteria of the genus Pseudomonas in the family Pseudomonadaceae of phylum Pseudomonadota.
- P. aeruginosa is an opportunistic nosocomial pathogen causing serious infection of the respiratory tract, urinary tract, blood, and wounds. It is the most common pathogen causing infection of burn wounds and outer ears.
- Multidrug-resistant strains of P. aeruginosa are increasing globally. Most of them have resisted traditionally used antibiotics. Infection by such multi-drug resistant strains can have a mortality rate of up to 60%. Resistant to ciprofloxacin and levofloxacin have made it to be listed under the serious threat list of the CDC.
6. Enterobacter species
- Enterobacter is a genus of Gram-negative, facultatively anaerobic, lactose-fermenting, rod-shaped Gammaproteobacteria of the family Enterobacteriaceae in phylum Pseudomonadota.
- Enterobacter includes several pathogenic species which mainly cause opportunistic infection in immune-compromised patients. E. aerogenes, E. cloacae, and E. sakazak are common human pathogens in the genus Enterobacter.
- Enterobacter spp. are commonly associated with UTIs and RTIs. Multidrug-resistant species are resistant to most of the β-lactams and cephalosporins, which makes them very hard to treat.
Clinical Characteristics of ESKAPE Pathogens
ESKAPE has the biggest impact on hospital-acquired infections, accounting for about 2/3rd of the total nosocomial infections worldwide. The US CDC has estimated around 2 million nosocomial infections with 23,000 fatalities by the ESKAPE in the US alone, and the situation will worsen yearly.
ESKAPE pathogens are responsible for the following types of infections in hospitalized patients:
- Hospital Acquired Pneumonia
- Urinary Tract Infections
- Wound and Skin Infections
- Infection in the Site of Surgery
- Bacteremia and Endocarditis
- Soft Tissue Infections
ESKAPE Pathogens Epidemiology
ESKAPE pathogens are reported globally, mostly in hospital settings. Their prevalence rate is in increasing trend. Due to the lack of a proper surveillance system, no exact data is published (as of 2021), but results published by CDCs in the US and by other European researchers show their prevalence is increasing rapidly. S. aureus is responsible for the highest number of infections among the ESKAPE pathogens.
Antimicrobial Resistance in ESKAPE Pathogens
ESKAPE pathogens are multi-drug resistant (resistant to more than three classes of antibiotics). They are resistant to most of the antibiotics that were traditionally used to treat these bacteria.
ESKAPE pathogens have developed resistance to all or most antibiotics in classes like β-lactams, β-Lactamase –inhibitors, macrolides, ciprofloxacin, tetracyclines, lipopeptides, quinolones and fluoroquinolones, and oxazolidinones, and even the last line of defense like most of the polymyxins, glycopeptides, and carbapenems.
| Bacteria | Most Common Resistant Antibiotic | Most Serious Strain | Available Treatment Option |
| Enterococcus faecium | Vancomycin ampicillin cephalosporins linezolid teicoplanin piperacillin amikacin amoxicillin-clavulanate ampicillin- sulbactam tobramycin nalidixic-acid imipenem meropenem | Vancomycin-resistant E. faecium | Nitrofurantoin Fosfomycin Chloramphenicol Daptomycin Doxycycline Omadacycline high-dosage ampicillin-sulbactam |
| Staphylococcus aureus | β-lactams Aminoglycosides Macrolides Chloramphenicol Trimethoprim Tetracyclines Tobramycin Cephatholin Carbapenem Piperacillin Piperacillin-tazobactam TicarcillinOxacillin | Methicillin-resistant S. aureus (MRSA) | Vancomycin Clindamycin Daptomycin Linezolid Dalbavancin Tigecycline Pristinamycin Trimethoprim-sulfamethoxazole Lefamulin Telavancin |
| K. pneumoniae | Polymyxins Carbapenems Fluoroquinolones Cephalosporins β-lactams Aminoglycosides Tetracyclines Nalidixic acid Ampicillin Amoxiclav Ticarcillin Cephalothin | Carbapenem resistant K. pneumoniae (CRKP) | Carbapenem-polymyxins combination Aminoglycosides Imipenem-cilastatin-relebactam Ceftazidime-avibactam Plazomicin |
| A. baumannii | Polymyxins Carbapenems β-lactams Fluoroquinolones Cephalosporins Tigecycline Ceftazidime Mezlocillin Ticarcillin | Carbapenem resistant A. baumannii (CRAB) | Colistin Carbapenem-polymyxin combination High dosage carbapenem Cefiderocol Eravacycline |
| P. aeruginosa | Ciprofloxacin Levofloxacin Cephalosporins Piperacillin-tazobactam Carbapenems Aminoglycosides Quinolones Polymyxins Cefoperazone Moxalactam Colistin Ticarcillin Ticarcillin-clavulanate Penicillins Ampicillin Amoxicillin Most β-lactams | Ciprofloxacin resistant P. aeruginosa | Imipenem-cilastatin-relebactam combination Piperacillin-tazobactam Ceftolozane-tazobactam Meropenem Ceftazidime-avibactam Cefiderocol |
| Enterobacter spp. | β-lactams Carbapenems Cephalosporins Fluoroquinolones Polymyxins Nalidixic acid | Pandrug-resistant Enterobacter spp. | Nitrofurantoin Cefepime Aztreonam Ceftriaxone Ciprofloxacin Gentamycin Meropenem Piperacillin-tazobactam Trimethoprim Imipenem-cilastatin-relebactam |
**Note: Not all strains are resistant or sensitive to antibiotics mentioned above. These apply to the most common resistant clinical species isolates.
References
- De Oliveira, M. P., Forde, B. M., Kidd, T. J., Harris, N. A., Schembri, M. A., Beatson, S. A., Paterson, D. L., & Walker, M. J. (2020). Antimicrobial Resistance in ESKAPE Pathogens. Clinical Microbiology Reviews, 33(3). https://doi.org/10.1128/CMR.00181-19
- Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., & Pardesi, K. R. (2019). Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Frontiers in Microbiology, 10. https://doi.org/10.3389/fmicb.2019.00539
Mechanism of Resistance in ESKAPE Pathogens
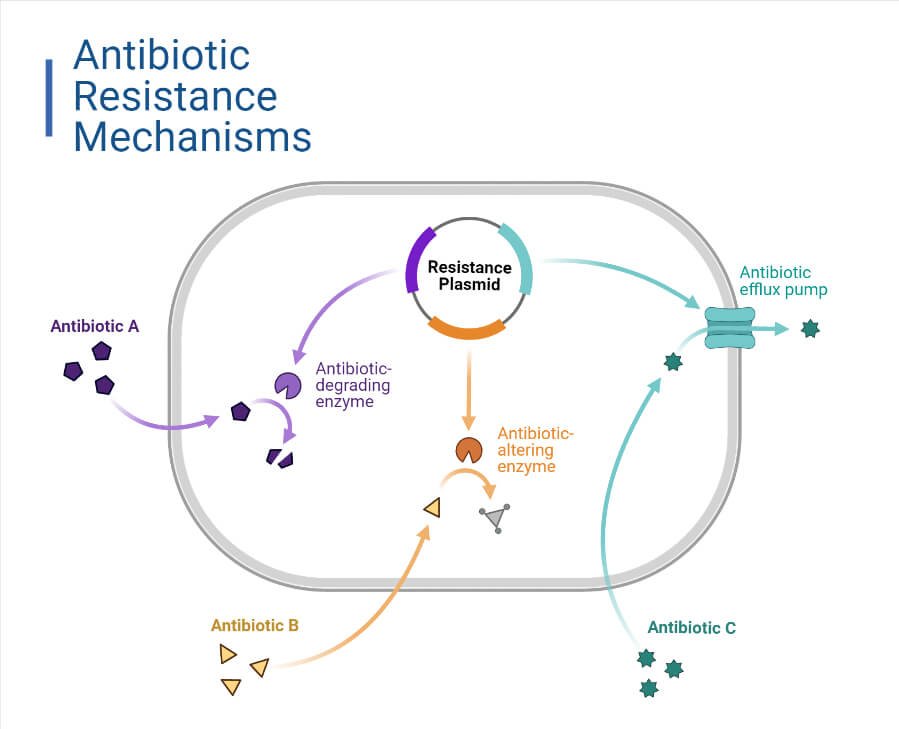
ESKAPE pathogens have developed several resistance mechanisms to escape the action of different classes of antibiotics. Some of the most common mechanisms are:
1. Antimicrobial Inactivation or Alteration
- The production of enzymes that irreversibly destroy or modify the antimicrobial molecules resulting in their inactivation or alteration, is the most common antimicrobial resistant (AMR) mechanism.
- The Gram-negative bacteria in the ESKAPE group (Pseudomonas, Acinetobacter, Klebsiella, Enterobacter) produce enzymes that destroy active sites of the antimicrobials or modify the main structural components of the antimicrobial.
- β- Lactamases: β-Lactamases enzyme production is the main AMR mechanism in Gram-negative ESKAPE pathogens. (Staphylococcus and Enterococcus are also reported to produce penicillinases and a few other β-Lactamases). These enzymes confer resistance against the – Lactam antibiotics by hydrolyzing the β-Lactam ring of the antibiotics. Extended-spectrum β-Lactamases (ESBLs), AmpC β-Lactamases (ABLs), Carbapenemases, and Metallo-β-lactamases (MBLs) are the most important clinically important β-Lactamases produced by ESKAPE pathogens. Production of these enzymes results in resistance to antibiotics like penicillins, cephalosporins, carbapenems, and monobactams.
- Aminoglycoside Modifying Enzymes (AMEs): These enzymes account for the most resistance against aminoglycoside antibiotics in the ESKAPE pathogens. These enzymes covalently catalyze the modification of specific amino groups or hydroxyl groups of aminoglycosides and reduce their affinity to bind with bacterial ribosomes. Aminoglycoside acetyl-transferases (AACs), aminoglycoside phosphor-transferases (APHs), and aminoglycoside nucleotidyltransferase (ANTs) are the three classes of AMEs. Among them, AACs are the main enzymes produced by ESKAPE pathogens. AMEs help ESKAPE pathogens to escape from the actions of amikacin, tobramycin, neomycin, plazomicin, gentamycin, etc.
2. Antimicrobials’ Target Modification
- Modifying the antibiotic target sites is another common AMR mechanism in ESKAPE pathogens to prevent themselves from the deleterious effect of antibiotics. Modifying the target sites reduces the affinity of antibiotic molecules to bind with the bacterial components.
- Bacteria can modify target proteins, enzymes, ribosomes, cell wall components, nucleic acid binding sites, etc., to develop resistance against antibiotics.
- Modification of Target Enzymes/Protein: It is a common method to escape from the action of β-Lactams and other antibiotics that target specific proteins. Bacteria modify Penicillin Binding Proteins (PBPs), which are the target site of the β-Lactams. Similarly, modification in DNA gyrase and Topoisomerase IV enzymes helps bacteria to prevent themselves from the action of quinolones like Nalidixic acid and fluoroquinolones like ciprofloxacin, ofloxacin, norfloxacin, etc.
- Modification in Ribosomal Target Sites: Methylation of A2058 residue of 23S rRNA of 50S ribosomal subunit of bacterial ribosomes confers resistance against macrolides, lincosamide, and streptogramin antibiotics. It is seen mainly in S. aureus and Enterococcus spp. This is the major AMR mechanism providing resistance against antibiotics like azithromycin, erythromycin, clarithromycin, etc.
Methylation of A2503 reside of 23S rRNA of 50S ribosomal subunit contribute resistance to linezolid in S. aureus and Enterococcus spp.
Also, methylation of 16S rRNA residue decreases the affinity of aminoglycoside binding to the bacterial ribosome.
- Modification in Cell-wall Precursors: It is one of the most important AMR mechanisms in S. aureus and E. faecium which provides resistance against glycopeptides to these Gram-positive ESKAPE pathogens. These pathogens synthesize modified peptidoglycan precursors (D – Ala – D – Lactate or D – Ala – D – Serine in place of traditional D – Ala – D – Ala) that reduce the glycopeptides binding. Additionally, they produce D, D – carboxypeptidases enzymes that eliminate remaining D – Ala – D – Ala. This causes modification in cell-wall precursors and cell walls of the pathogens, hence developing resistance against glycopeptides like vancomycin, daptomycin, oritavancin, etc.
3. Modification in Membrane Permeability
- Reduction in membrane permeability causes a reduction in uptake of antibiotics; hence the concentration of intracellular antibiotics will be a low resulting decrease in the action of antibiotics.
- One of the main mechanisms to reduce membrane permeability is a modification in porins or down-regulation or loss of porins channels in the outer membrane. Modification in porins contributes to resistance to β-Lactams and fluoroquinolones, which are porin dependent on penetrating the bacterial cells. It is a major AMR mechanism in carbapenem resistance Acinetobacter baumannii and Pseudomonas aeruginosa.
4. Efflux Pumps
- Efflux pumps that extrude intracellular antibiotics contribute greatly to the AMR mechanism. There are six classes of efflux pumps in bacteria related to AMR, and all of these are found in ESKAPE pathogens; they are resistance-nodulation-division (RND), major facilitator superfamily (MFS), multidrug and toxic compound extrusion (MATE), small multidrug resistance (SMR), ATP-binding cassette (ABC), and proteobacterial antimicrobial compound efflux (PACE). Most of these efflux pumps are responsible for effusing different types of antibiotics rather than those in a single category.
- RND type efflux pump plays a major role in conferring multi-drug resistance to Gram-negative ESKAPE pathogens. They expel not only antibiotics but also dyes and detergents. MexAB-OprM and MexCD-OprJ are two important efflux pumps of RND types that develop resistance against aminoglycosides, fluoroquinolones, and carbapenems.
- AcrAB efflux pump is responsible for developing resistance against imipenem, tetracycline, chloramphenicol, and fluoroquinolones in Enterobacter aerogenes and K. pneumoniae.
- Multidrug-resistant strains of P. aeruginosa and A. baumannii show an increase in AdeABC, AdeDE, AdeFGH, and AdeIJK efflux pumps of the RND family. This increases resistance against chloramphenicol, aminoglycosides, fluoroquinolones, tetracycline, and erythromycin.
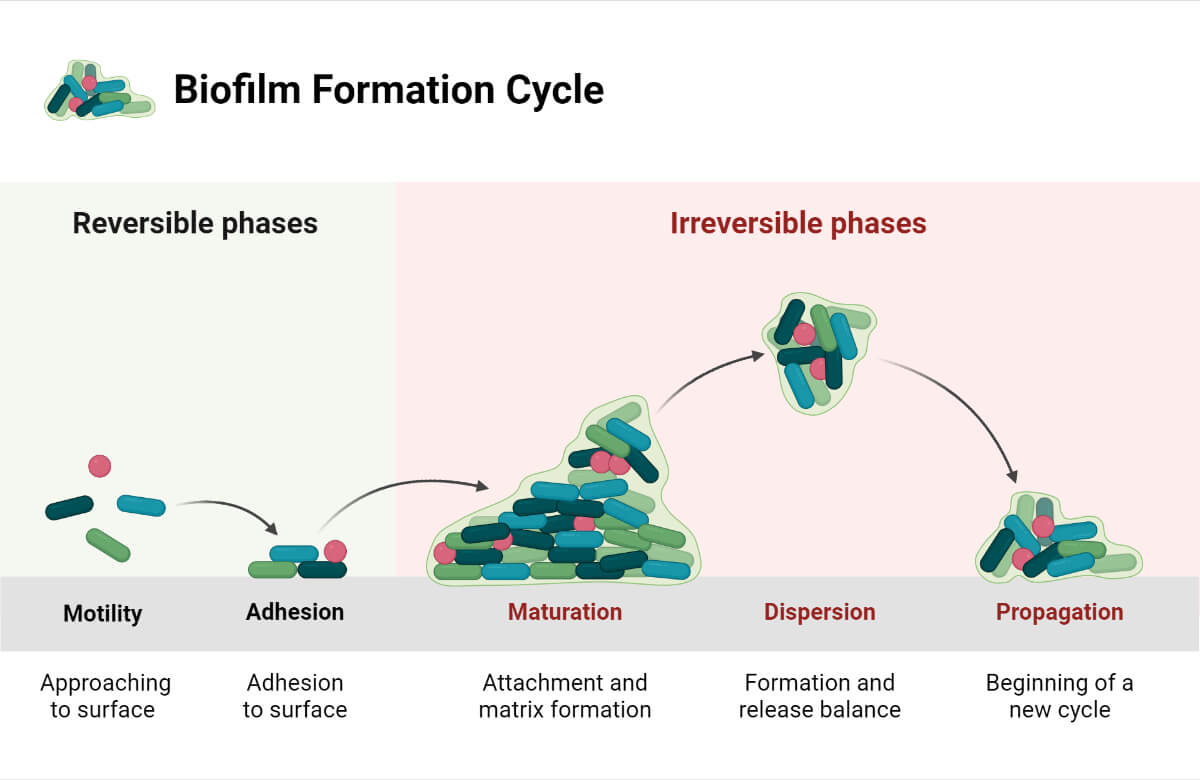
5. Biofilm Formation by ESKAPE Pathogens
- Biofilm formation is an important phenomenon demonstrated mainly by S. aureus, P. aeruginosa, A. baumannii, and K. pneumoniae among the ESKAPE pathogens. Biofilm not only increases the chances of survival in medical devices (or inanimate surfaces) but also provides resistance to antibiotics.
- Biofilm increases resistance against antibiotics by restricting antibiotic penetration, modifying antibiotics, increasing interaction between different bacterial species, upregulating effluxes, enhancing horizontal gene transfer, etc.
Works being done against ESKAPE Pathogens
- WHO, CDC, and ECDC have listed ESKAPE pathogens under urgent and serious threat lists and have prioritized their surveillance, research, and focus on them. A major focus is research and development of new effective antimicrobials, antimicrobial stewardship, and prevention of hospital-acquired infections (HIAs).
- There is limited treatment option left to fight against ESKAPE pathogens. Besides the treatment mentioned above (in the table), several drugs are under research (as of 2020) and trials; like combination therapy, silver nano-particle therapy, phage therapy, and new antibiotics like plazomicin, cefiderocol, meropenem – vaborbactam, imipenem + cilastatin + relebactam, sulbactam – durlobactam, ceftobiprole, tebipenem – pivoxil, lascufloxacin, etc.
- Study on lytic phage against ESKAPE pathogens in hospital wastewater.
- CLSI annually updates the antibiotic sensitivity testing standards and lists of antibiotics to be employed against ESKAPE pathogens.
References
- Drug Resistance – There is No Escape from the ESKAPE Pathogens (emerypharma.com)
- 2019 Antibiotic Resistance Threats Report | CDC
- The ESKAPE bacteria group and its clinical importance – Clover Bioanalytical Software (cloverbiosoft.com)
- ESKAPES: Emerging Pathogens of Concern (infectioncontrol.tips)
- J_Paramed_Sci_2016_7_3_43_57.pdf (who.int)
- Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., & Pardesi, K. R. (2019). Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Frontiers in Microbiology, 10. https://doi.org/10.3389/fmicb.2019.00539
- De Oliveira, M. P., Forde, B. M., Kidd, T. J., Harris, N. A., Schembri, M. A., Beatson, S. A., Paterson, D. L., & Walker, M. J. (2020). Antimicrobial Resistance in ESKAPE Pathogens. Clinical Microbiology Reviews, 33(3). https://doi.org/10.1128/CMR.00181-19
- Santajit, S., & Indrawattana, N. (2016). Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Research International, 2016. https://doi.org/10.1155/2016/2475067
- Arbune, M., Gurau, G., Niculet, E., Iancu, A. V., Lupasteanu, G., Fotea, S., Vasile, M. C., & Tatu, A. L. (2021). Prevalence of Antibiotic Resistance of ESKAPE Pathogens Over Five Years in an Infectious Diseases Hospital from South-East of Romania. Infection and Drug Resistance, 14, 2369-2378. https://doi.org/10.2147/IDR.S312231
- Pendleton, Jack N; Gorman, Sean P; Gilmore, Brendan F (2013). Clinical relevance of the ESKAPE pathogens. Expert Review of Anti-infective Therapy, 11(3), 297–308. doi:10.1586/eri.13.12
- Schultz, F., Anywar, G., Tang, H. et al. Targeting ESKAPE pathogens with anti-infective medicinal plants from the Greater Mpigi region in Uganda. Sci Rep 10, 11935 (2020). https://doi.org/10.1038/s41598-020-67572-8
