Pseudomonas putida is a Gram-negative flagellated rod-shaped bacterium that is commonly isolated from diverse environmental and clinical samples.
These organisms are frequent inhabitants of the rhizospheric and freshwater habitats and have diverse metabolic versatility against a wide range of biogenic and xenobiotic compounds.
P. putida is an uncommon cause of infections of various body sites, especially in immune-compromised individuals like newborns and cancer patients. P. putida is a part of the fluorescent group of Pseudomonas species as they produce different forms of fluorescent pigments. The most important characteristic that distinguishes P. putida from P. aeruginosa is the inability of P. putida to hydrolyze gelatin.
Besides, other features like the inability to produce any phenazine pigments, to denitrify, and to grow at 41°C also help in the differentiation of P. putida from P. fluorescens and P. aeruginosa species.
The genus name ‘putida’ is derived from the Latin term ‘putida’, meaning stinking or fetid, indicating the occurrence of the bacteria in spoiled food items and the aromatic odor on solid media. The species was discovered by Trevisan in 1889. Pseudomonas putida is considered an evolutionary group that includes other species like P. fulva, P. parafulva, P. alkylphenolia and P. monteilii.
Like most Pseudomonas species, P. putida also exhibits some form of antibiotic resistance against certain antibiotics as a result of the presence of plasmids bearing the genes that encode antibiotic resistance. The presence of such plasmids also increases the chances of the transfer of plasmids to other microorganisms in hospital environments.
P. putida is the first patented microorganism in the world because of its ability to decompose hydrocarbons. The diverse metabolism of the organism has been exploited for its use in bioremediation and industries. Similarly, the ability to produce secondary metabolites also enables the use of P. putida as a biocontrol agent to protect against pathogenic microorganisms.
Interesting Science Videos
Classification of Pseudomonas putida
- The classification of bacteria into different orders, families, and genus is crucial in the fields of environmental microbiology and epidemiology.
- Traditionally, the classification was made on the basis of phenotypic and metabolic characteristics of the organisms like morphology, cultural characteristics, and biochemical testing.
- More recently, the basis of classification has been changed to the analysis of the nucleic acid components.
- The most common basis for the classification of bacteria includes DNA sequences via methods like DNA fragment analysis and DNA sequence analysis.
- At the genus and species level, however, 16S rRNA gene sequence analysis is adopted.
- Pseudomonas putida is considered an evolutionary group and includes several other species based on the common 16S rRNA sequences.
- P. putida is further classified into two biovars; biovar A and biovar B. there are about 103 strains in biovar A and about nine strains belonging to biovar B.
The following is the taxonomical classification of P. putida:
| Domain | Bacteria |
| Phylum | Proteobacteria |
| Class | Gammaproteobacteria |
| Order | Pseudomonadales |
| Family | Pseudomonadaceae |
| Genus | Pseudomonas |
| Species | P. putida |
Habitat of Pseudomonas putida
- Pseudomonas putida is a saprophyte that can be found in various ecological habitats like soil (mostly rhizosphere), drains, fresh and saltwater, and animate surfaces of plants, animals, and human beings.
- The ability of an organism to survive in different habitats defer on seasons and the adaptive mechanism adopted by the organism.
- These organisms have extraordinarily broad niches as a result of their metabolic and competitive capabilities.
- They can use a variety of metabolic compounds and can withstand large ranges of abiotic challenges.
- Even though, P. putida and P. fluorescens tend to be present in similar habitats, the occurrence of P. putida is more in household sites which is due to the occurrence of P. putida in sewage and wastewater.
- P. putida is not isolated from most high-temperature sites that can harbor P. fluorescens and P. aeruginosa due to the inability of P. putida to survive at higher temperatures like 42°C.
- Most strains of P. putida have a broad global distribution with some species being isolated from soil samples in and around lakes in Antarctica.
- However, there are some strains that have a more restricted distribution as a result of a less versatile metabolism.
- In animals and humans, P. putida occurs as non-pathogenic species, but some strains can act as opportunistic pathogens in immunocompromised individuals and newborns.
- P. putida are obligate aerobes and thus, require oxygen-rich environments for growth and development.
- Some members of the P. putida group belong to the normal inhabitants of plant parts like roots and leaves.
Morphology of Pseudomonas putida
- The cells of P. putida are Gram-negative rod-shaped cells with a size of 0.5 to 1.0 µm in length.
- The length of the cells might differ depending on the age of the cell on culture media as young cells tend to be longer.
- Some cells of the organism might have a variable width to length ratio depending on the culture conditions. Some cells are longer and broader and might look filamentous.
- Most cells occur as single cells, but some pairing might be observed in an end-to-front fashion.
- The outer surface of the cells consists of flagella as P. putida is a motile bacterium. The number of flagella can be either one or more.
- In addition to the flagella, cells also consist of fimbriae or pili on the outer surface, which helps in movement as well as attachment.
- The cell envelope forms the outermost covering of the bacteria, which is composed of three distinct layers; an outer membrane, a peptidoglycan layer, and a cell membrane.
- The peptidoglycan layer is responsible for protecting the bacteria against other microbes and antibacterial agents.
- The cell membrane is made up of a lipid bilayer, which is equipped to change the fluidity of the membrane according to the outer environmental conditions.
- The number of proteins in P. putida ranges between 3748-6780, whereas the GC content of the genome is about 60%.
- Pseudomonas putida belong to the fluorescent group of Pseudomonas species and produce a fluorescent pigment pyoverdine. They do not, however, produce phenazine pigments.
Cultural Characteristics of Pseudomonas putida
- Pseudomonas putida isolated from environmental samples are quite easy to grow on artificial culture media as they have minimum nutritional requirements.
- The temperature of growth for P. putida is within the range of 15-35°C, and the optimum temperature for growth is between 30-35°C.
- P. putida can be distinguished from other fluorescent Pseudomonas based on the inability of the organism to survive at 41°C.
- Similarly, the optimum pH for growth lies between pH values of 7-8. The bacteria flourish in the presence of oxygen.
- Depending on the samples, five different types of colonies of P. putida can be seen in artificial culture media.
- Environmental isolates are circular and smooth that are convex, and dome-shaped with a shining surface.
- Clinical isolates tend to produce mucoid colonies with entire edges that appear opaque, homogenous, whitish, or greyish-green in color.
- Colonies of fluorescent Pseudomonas like P. putida usually form wrinkly spreader and fuzzy spreader type colonies on artificial media.
- The following are some cultural characteristics of P. putida in different culture media:
1. Pseudomonas putida in Nutrient Agar
- Smooth white colonies with entire edges and convex shapes are observed on NA.
- The younger colonies appear circular, and they might get wider after longer incubation. Besides, the colonies are butyrous, glistening, nearly opaque, and pale green with darkened centers.
- Under sunlight or UV radiation, the colonies produce a greenish color which can dissolve in the media.
- The environmental samples of P. putida from soil and water are usually isolated on NA.
2. Pseudomonas putida in Cetrimide Agar
- The growth of bacteria on Cetrimide agar is taken as a test for the identification of Pseudomonas species as the agar is selective for Pseudomonas species.
- The colonies on the cetrimide agar appear dry and flat with a circular circumference and lobular edges.
- On the cetrimide agar, opaque and greenish colonies are seen as a result of the diffusion of pigment onto the culture media.
- The growth of P. putida on Cetrimide agar is often performed as a form of identification of the organism.
3. Pseudomonas putida in MacConkey Agar
- On MacConkey agar, colonies of P. putida appear round and flat. The color of the media remains unchanged as the organism is a lactose non-fermenter.
- The organism does, however, produce a pigment that diffuses through the media, resulting in a greenish coloration around the colonies.
Biochemical Characteristics of Pseudomonas putida
The biochemical characteristics of P. putida can be tabulated as follows:
| S.N | Biochemical Characteristics | Pseudomonas putida |
| 1. | Capsule | Non-Capsulated |
| 2. | Shape | Rod |
| 3. | Gram Staining | Gram-Negative |
| 4. | Catalase | Positive (+) |
| 5. | Oxidase | Positive (+) |
| 6. | Citrate | Negative (-) |
| 7. | Methyl Red (MR) | Negative (-) |
| 8. | Voges Proskauer (VR) | Negative (-) |
| 9. | OF (Oxidative-Fermentative) | Oxidative |
| 10. | Coagulase | Negative (-) |
| 11. | DNase | Negative (-) |
| 12. | Urease | Negative (-) |
| 13. | Gas | Negative (-) |
| 14. | H2S | Negative (-) |
| 15. | Hemolysis | β-hemolytic |
| 16. | Motility | Motile with multiple flagella |
| 17. | Nitrate Reduction | Negative (-) |
| 18. | Gelatin Hydrolysis | Negative (-) |
| 19. | Starch Hydrolysis | Negative (-) |
| 20. | Casein Hydrolysis | Negative (-) |
| 21. | Pigment Production | Positive (+) (Yellow-green) |
| 22. | Indole | Negative (-) |
| 23. | TSIA (Triple Sugar Iron Agar) | Alkali/Alkali (Red/ Red) |
| 24. | Spore | Non-sporing |
| 25. | Cetrimide Test | Positive (+) |
| 26. | Denitrification | Negative (-) |
Fermentation
| S.N | Substrate | Pseudomonas putida |
| 1. | Adonitol | Negative (-) |
| 2. | Arabinose | Negative (-) |
| 3. | Cellobiose | Negative (-) |
| 4. | Dulcitol | Negative (-) |
| 5. | Fructose | Positive (+) |
| 6. | Galactose | Positive (+) |
| 7. | Glucose | Positive (+) |
| 8. | Glycerol | Positive (+) |
| 9. | Glycogen | Negative (-) |
| 10. | Hippurate | Positive (+) |
| 11. | Inulin | Negative (-) |
| 12. | Inositol | Negative (-) |
| 13. | Lactose | Negative (-) |
| 14. | Malonate | Positive (+) |
| 15. | Maltose | Negative (-) |
| 16. | Mannitol | Positive (+) |
| 17. | Mannose | Positive (+) |
| 18. | Pyruvate | Negative (-) |
| 19. | Raffinose | Negative (-) |
| 20. | Rhamnose | Negative (-) |
| 21. | Ribose | Positive (+) |
| 22. | Salicin | Negative (-) |
| 23. | Sorbitol | Negative (-) |
| 24. | Starch | Negative (-) |
| 25. | Sucrose | Negative (-) |
| 26. | Trehalose | Negative (-) |
| 27 | Xylose | Positive (+) |
Enzymatic Reactions
| S.N | Enzymes | Pseudomonas putida |
| 1. | Acetoin | Negative (-) |
| 2. | Acetate Utilization | Positive (+) |
| 3. | Arginine Dehydrolase | Positive (+) |
| 4. | Esculin Hydrolysis | Negative (-) |
| 5. | Lecithinase | Negative (-) |
| 6. | Lipase | Positive (+) |
| 7. | Lysine Decarboxylase | Negative (-) |
| 8. | Ornithine Decarboxylase | Negative (-) |
| 9. | Phenylalanine Deaminase | Negative (-) |
Virulence Factors of Pseudomonas putida
- Pseudomonas putida is a non-pathogenic bacterium that is found in environmental sources like air, water, and soil.
- The organism colonizes different parts of the animal body as an indigenous bacteria, but can rarely cause nosocomial infections in immunocompromised individuals.
- The most important attribute is the ability of P. putida to cause infections is its ability to survive at 37°C.
- It can also withstand a low temperature of 4°C and thus can survive in blood and other clinical samples stored for further use.
- There are different attributes and structures that support the mechanism of disease production by P. putida.
- P. putida is not as virulent as other fluorescent Pseudomonas species like P. aeruginosa and P. fluorescens, but it does, however, has some common factors like antibiotic resistance.
- The exact mechanism of disease or pathogenesis of P. putida is not yet understood, but it is known that antibiotic resistance and biofilm formation play an important part in the process.
Some of the common virulence factors associated with infections caused by P. putida can be described as follows:
1. Flagella and pili
- Flagella and pili represent the structural component of P. putida involved in the attachment and movement of bacteria through the tissues.
- P. putida has more than one flagella that enable the movement of bacteria via swimming motion.
- The flagella are driven by a motor present at the base which moves the filament in a helical way.
- Similarly, the pili present on the outer surface of the bacteria help in the binding of bacteria to the host cell surface.
- Both of these structures are crucial in the movement and colonization of the host tissue surface by P. putida.
- In addition to helping in colonization, flagella and pili are also important in the formation of biofilm by enabling the binding of several cells to one another.
2. Quorum Sensing and Biofilm
- Quorum sensing is an important mechanism of regulating various cells of the Pseudomonas population to assist the process of disease progression.
- Quorum sensing is responsible for the production of alginates that facilitate the accumulation of extracellular material and cells to form biofilms on animate and inanimate surfaces.
- One of the predisposing factors of biofilm formation is the presence of medical devices like catheters and respirators.
- The formation of biofilm helps in the protection of bacteria against the host immune system as well as antibiotic components.
3. Secondary metabolites
- Pseudomonas putida is among the few fluorescent species of Pseudomonas that are capable of producing secondary metabolites with antibacterial properties.
- These products include compounds like hydrogen cyanide and rhizoxin, which help the bacteria compete with other microbial agents.
- Hydrogen cyanide is crucial to fight against plant pathogens as it disrupts the intake of water and minerals by the competing microorganism.
- In the case of mammalian cells, the production of hydrogen cyanide by P. putida is regulated by hcnABC genes which are upregulated in the absence of oxygen.
4. Lipopolysaccharides
- The outer cell envelope of P. putida is composed of lipopolysaccharides that protect the bacteria against phagocytosis and lysis.
- Besides, the lipid A present on the membrane also induces cytotoxic activity and further assists protection against phagocytosis.
- The lipopolysaccharides also cause excessive stimulation of the immune system, resulting in the release of factors like IL-8 and TNF-α.
- It has been discovered that excessive inflammatory response is the primary cause of disease production in different organs.
5. Proteases
- The production of proteases like elastases and phospholipases are also involved in tissue damage and facilitating the dissemination of bacteria to different areas.
- Phospholipase is involved in the degradation of phospholipids which form an important constituent of the lipid component of the body.
- One of the products of phospholipid degradation is diacylglycerol which has toxigenic activity against host cells by inducing the production of metabolites like protein kinase and arachidonic acid.
- These metabolites further induce an inflammatory response and cause tissue damage.
Pathogenesis of Pseudomonas putida
- Since most strains of P. putida are not associated with diseases and are saprophytes found in the natural environment, much is not known about the pathogenesis of disease caused by P. putida.
- It is known that infections occur in immunocompromised individuals as a result of induced inflammatory response.
- Common sites of infection include the lungs and the bloodstream, which if disseminated might cause severe diseases.
- The occurrence of P. putida infections is more common in patients with cystic fibrosis and AIDS as the immune response in such patients is either slow or reduced.
- The entry of P. putida into the human body can occur either via transfusion of contaminated blood or as a result of the movement of bacteria into the sterile sites of the body.
- The overall process of the pathogenesis of P. putida is facilitated by various virulence factors that help in entry, colonization, tissue damage, and antibiotic resistance.
The process of the pathogenesis of infections caused by P. putida can be explained below:
1. Entry and Colonization
- The entry of the bacteria into the host body can occur either due to the transfusion of contaminated blood in the hospital setting or due to the entry of the bacteria to the sterile sites of the body through wounds or burns.
- Flagella and pili are two crucial structures that facilitate the entry of bacteria into the host tissue. Flagella enable the movement of bacteria while pili facilitate the attachment of bacteria to the target sites.
- During colonization, the bacteria escape phagocytosis as the lipopolysaccharide layer of the bacteria protects against the activity of macrophages.
- Once attached to the host cell surface, the bacteria start taking minerals and nutrients from the host cell in order to survive and reproduce.
2. Tissue damage
- The second step in the pathogenesis of P. putida infections is tissue damage and dissemination of bacteria to different parts of the body.
- This is facilitated by the production of various proteins like enzymes and toxins that help degrade the host cell.
- Some of the common enzymes involved in the process include proteases like elastase which degrades the protein elastin and also enables the dissemination of bacteria to the neighboring cells.
- Phospholipase, on the other hand, degrades the phospholipids present on the cell membrane of the host cell and causes the release of cytoplasmic components, eventually leading to necrosis.
- The products of this degradation can induce an inflammatory response by the host immune system, which causes further tissue damage.
3. Biofilm formation
- Biofilm formation is the method of protection against the host immune response as well as against antimicrobial agents.
- The biofilm formation in P. putida is influenced by compounds by alginates and the extracellular matrix produced by the bacteria.
- The biofilm provides a protective environment which helps in moisture retention and availability of nutrients to all the bacterial cells.
- In addition to metabolic products involved in biofilm formation, structural components like pili also play an essential role.
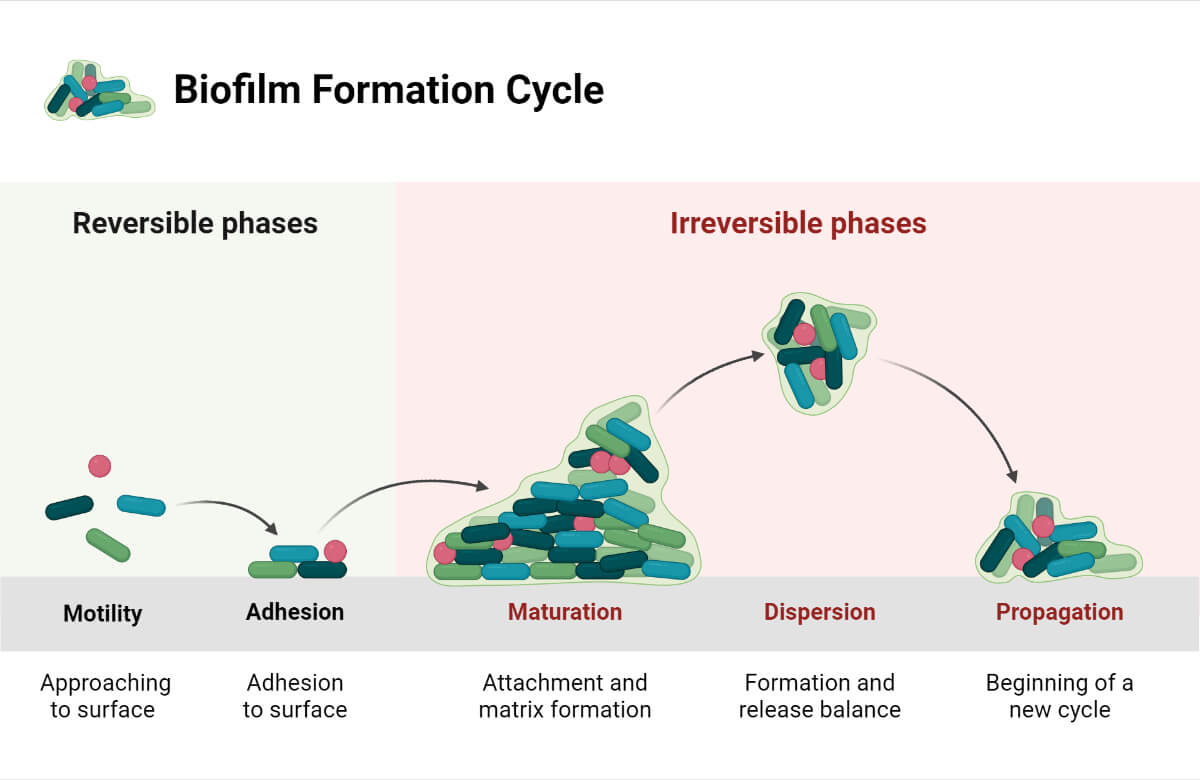
Clinical Manifestation of Pseudomonas putida
- Infections caused by P. putida are specific to particular sites in the body where the bacteria colonizes and reproduces.
- Some of the common sites of infections caused by P. putida include the bloodstream, soft tissues, and lungs.
- Most of these infections are mild; however, they might become severe if the patients have some underlying conditions.
- The underlying conditions can be either mucocutaneous defect or underlying compromised immunity.
- The infections can result in fulminant sepsis-induced multiorgan failure, leading to death.
Some of the common clinical manifestations of infections caused by P. putida include:
1. Bacteremia
- Bacteremia is the most dreadful and common manifestation of P. putida, where the bacteria reaches the bloodstream of the patients.
- Periodic fever and chills are observed as a new set of bacteria are released into the bloodstream after reproduction.
- These infections can either occur after soft tissue infections as the bacteria makes its way into the bloodstream or due to the transfusion of contaminated blood.
- There have been cases of bacteremia that further develop into fulminant sepsis-induced multiorgan failure.
- The severity of infections is more common in patients with some underlying conditions.
- Other factors like advanced age and malnutrition are possible causes of the reduced immune response despite aggressive treatment.
2. Soft-tissue infections
- Skin and soft tissue infections caused by P. putida are uncommon and are usually only observed in immunocompromised individuals.
- A common site of infections includes cartilages and connective tissues. The infections can be detected through symptoms like the formation of ulcerative lesions in the target sites.
- The entry of bacteria is assumed to be through trauma on the skin surface which enables the bacteria to reach the target sites.
- Most patients tend to have some underlying conditions, like HIV infection and renal insufficiency.
- In the absence of appropriate antimicrobial coverage, the condition of the patient might worsen, which can lead to bacteremia and death.
3. Respiratory infections
- Instances of hospital-acquired respiratory infections are also possible outcomes of P. putida infections.
- The source of bacteria is assumed to be contaminated bronchoscopes which transmit the infection from one patient to another in hospital settings.
- The isolation of bacteria from samples like the tracheal aspirates and sputum can be used as a method of diagnosis of P. putida infection as it can be confused with other infections otherwise.
Lab Diagnosis of Pseudomonas putida
- Laboratory diagnosis of bacterial infections is often associated with the isolation and identification of bacteria from the site of infections.
- The process of diagnosis also depends on the type of sample used. In the case of blood, culture is performed to isolate bacteria.
- The preliminary stage of identification is based on the isolation and identification of bacteria based on their morphological and biochemical characteristics.
- Other more sophisticated methods, including immunological and molecular methods, provide more accurate identification and diagnosis.
- Early diagnosis of infections caused by P. putida is essential for early treatment as the progression of the disease might cause complications.
The following are the detailed methods of laboratory diagnosis of P. putida from clinical samples:
1. Preliminary identification
- The methods of preliminary identification of P. putida involve the isolation of bacteria from the clinical samples.
- Isolation is achieved by the growth of bacteria on an artificial growth media. The colony morphology produced by the bacteria provides some information about the identification of the bacteria.
- It is followed by the microscopic observation of the organism to observe its morphology. In order to confirm the presence of P. putida in the clinical sample, biochemical tests specific to P. putida are performed.
- Gelatin hydrolysis test and the ability to grow at 41°C are two of the important biochemical test to differentiate P. putida from other Pseudomonas species.
2. Immunological methods
- Immunoassays are based on the identification of proteins and enzymes that are specific to the bacteria via techniques like ELISA and Western Blotting.
- Besides, other rapid tests can also be performed in order to identify the bacteria based on the presence or absence of antigen and other similar structures.
3. Molecular methods
- The molecular method of disease diagnosis is a novel technique that is continuously being applied in modern medicine for a more reliable and accurate result.
- Methods like PCR and DNA sequencing are slowly replacing the preliminary diagnosis methods as those are more time-consuming.
- Molecular methods detect certain proteins, DNA segments, or rRNA fragments that are specific to a certain bacteria.
Treatment of Pseudomonas putida
- Ceftazidime is the drug of choice for P. putida along with imipenem and meropenem.
- Besides, other antibacterial drugs like carbapenems, aminoglycosides, tetracyclines, and polymyxin B can also be used as a possible treatment regime against P. putida.
- Antibiotic resistance is a problem with the treatment of P. putida as the bacteria is found to be resistant against carbenicillin and gentamicin, which are used for the treatment of P. aeruginosa.
- In the case of devices-related infection, removal of such devices is required. Depending on the severity of the disease, ICU admission might be required.
- Antibiotic susceptibility testing should be performed to determine the susceptibility of bacteria against different antibiotics.
Prevention of Pseudomonas putida
- Transmission of P. putida occurs in a hospital environment which indicates that prevention of diseases can be achieved by following proper guidelines in the hospital setting.
- Most bacteria are transmitted via hospital personnel during a blood transfusion, thus performing blood testing before a blood transfusion is essential to prevent such infections.
- The health workers should use gloves and coats to prevent the transfer of bacteria within patients or to them.
- The use of contaminated food or flowers should be avoided as such items can act as vehicles for the bacteria.
- Since the infections begin with topical infections of the skin, treatment of such sites should be done immediately to avoid further complications.
Industrial Uses / Applications of Pseudomonas putida
- One of the most important applications of P. putida is the use of this bacteria in bioremediation. The central routes of carbon metabolism in P. putida consist of various conversing pathways of different substrates which provide building blocks and cofactors of industrial importance.
- The versatile metabolism of the bacteria allows the use of bacteria in unfavorable conditions for the degradation of otherwise non-degradable compounds.
- The bacteria have also been used as soil inoculants to reduce the naphthalene contamination in soil.
- The bacteria can also convert styrene oil into biodegradable plastic PHA which is then acted upon by other microorganisms.
- Since many strains of P. putida can produce different antibiotic secondary metabolites, these can also be used as a form of biocontrol in agriculture.
- P. putida has also been isolated from rhizospheric regions of various plants indicating its possible application in agriculture as a plant growth-promoting rhizobacteria.
References
- Topley W. W. C (2007). Topley and Wison’s Microbiology and Microbial Interactions; Bacteriology, 2 Vol. Tenth Edition. John Wiley and Sons Ltd.
- Bergey, D. H., Whitman, W. B., De, V. P., Garrity, G. M., & Jones, D. (2009). Bergey’s manual of systematic bacteriology: Vol. 2. New York: Springer.
- Loeschcke, Anita, and Stephan Thies. “Pseudomonas putida-a versatile host for the production of natural products.” Applied microbiology and biotechnology vol. 99,15 (2015): 6197-214. doi:10.1007/s00253-015-6745-4
- Gianluigi Lombardi, Francesco Luzzaro, Jean-Denis Docquier, Maria Letizia Riccio, Mariagrazia Perilli, Alessandra Colì, Gianfranco Amicosante, Gian Maria Rossolini, Antonio Toniolo. Nosocomial Infections Caused by Multidrug-Resistant Isolates of Pseudomonas putida Producing VIM-1 Metallo-β-Lactamase. Journal of Clinical Microbiology Nov 2002, 40 (11) 4051-4055; DOI: 10.1128/JCM.40.11.4051-4055.2002
- Calb, R et al. “Structure and function of the Pseudomonas putida integration host factor.” Journal of bacteriology vol. 178,21 (1996): 6319-26. doi:10.1128/jb.178.21.6319-6326.1996
- Thomas, Benjamin S et al. “A Lethal Case of Pseudomonas putida Bacteremia Due to Soft Tissue Infection.” Infectious diseases in clinical practice (Baltimore, Md.) vol. 21,3 (2013): 147-213. doi:10.1097/IPC.0b013e318276956b
- Peter, S., Oberhettinger, P., Schuele, L. et al. Genomic characterization of clinical and environmental Pseudomonas putida group strains and determination of their role in the transfer of antimicrobial resistance genes to Pseudomonas aeruginosa . BMC Genomics 18, 859 (2017). https://doi.org/10.1186/s12864-017-4216-2
- Teramoto K, Sato H, Sun L, Torimura M, Tao H, Yoshikawa H, Hotta Y, Hosoda A, Tamura H. Phylogenetic classification of Pseudomonas putida strains by MALDI-MS using ribosomal subunit proteins as biomarkers. Anal Chem. 2007 Nov 15;79(22):8712-9. doi: 10.1021/ac701905r. Epub 2007 Oct 16. PMID: 17939647.
- Remold, S.K., Brown, C.K., Farris, J.E. et al. Differential Habitat Use and Niche Partitioning by Pseudomonas Species in Human Homes. Microb Ecol 62, 505 (2011). https://doi.org/10.1007/s00248-011-9844-5
- Kuddus, Mohammed & Joseph, Babu & Ramteke, Pramod. (2013). Production of laccase from newly isolated Pseudomonas putida and its application in bioremediation of synthetic dyes and industrial effluents. Biocatalysis and Agricultural Biotechnology. 2. 333–338. 10.1016/j.bcab.2013.06.002.
- Susse Kirkelund Hansen, Janus A. J. Haagensen, Morten Gjermansen, Thomas Martini Jørgensen, Tim Tolker-Nielsen, Søren Molin. Characterization of a Pseudomonas putida Rough Variant Evolved in a Mixed-Species Biofilm with Acinetobacter sp. Strain C6. Journal of Bacteriology Jun 2017, 189 (13) 4932-4943; DOI: 10.1128/JB.00041-07
- Gulez, Gamze et al. “Colony morphology and transcriptome profiling of Pseudomonas putida KT2440 and its mutants deficient in alginate or all EPS synthesis under controlled matric potentials.” MicrobiologyOpen vol. 3,4 (2014): 457-69. doi:10.1002/mbo3.180
- Maksimova NP, Blazhevich OV, Lysak VV, Fomichev IuK. Kharakteristika fluorestsiruiushchego pigmenta pioverdina Pm, produtsiruemogo bakteriiami Pseudomonas putida [Characterization of the fluorescent pigment pyoverdine Pm, produced by Pseudomonas putida bacteria]. Mikrobiologiia. 1994 Nov-Dec;63(6):1038-44. Russian. PMID: 7760765.
- Ramon Zulueta-Rodriguez, Miguel Victor Cordoba-Matson, Luis Guillermo Hernandez-Montiel, Bernardo Murillo-Amador, Edgar Rueda-Puente, Liliana Lara, “Effect of Pseudomonas putida on Growth and Anthocyanin Pigment in Two Poinsettia (Euphorbia pulcherrima) Cultivars”, The Scientific World Journal, vol. 2014, Article ID 810192, 6 pages, 2014. https://doi.org/10.1155/2014/810192
- Thomas, Benjamin S et al. “A Lethal Case of Pseudomonas putida Bacteremia Due to Soft Tissue Infection.” Infectious diseases in clinical practice (Baltimore, Md.) vol. 21,3 (2013): 147-213. doi:10.1097/IPC.0b013e318276956b
- Kim, Seong Eun et al. “Nosocomial Pseudomonas putida Bacteremia: High Rates of Carbapenem Resistance and Mortality.” Chonnam medical journal vol. 48,2 (2012): 91-5. doi:10.4068/cmj.2012.48.2.91
- Fernández, Matilde et al. “Analysis of the pathogenic potential of nosocomial Pseudomonas putida strains.” Frontiers in microbiology vol. 6 871. 25 Aug. 2015, doi:10.3389/fmicb.2015.00871
- Poblete-Castro, Ignacio & Becker, Judith & Dohnt, Katrin & Santos, Vitor & Wittmann, Christoph. (2012). Industrial biotechnology of Pseudomonas putida and related species. Applied microbiology and biotechnology. 93. 2279-90. 10.1007/s00253-012-3928-0.
