DNA sequencing refers to methods for determining the order of the nucleotide bases adenine,guanine,cytosine, and thymine in a molecule of DNA.
The first DNA sequence was obtained by academic researchers, using laboratories methods based on 2- dimensional chromatography in the early 1970s.
By the development of dye based sequencing method with automated analysis, DNA sequencing has become easier and faster.
Two main methods are widely known to be used to sequence DNA:
- The Chemical Method (also called the Maxam–Gilbert method after its inventors).
- The Chain Termination Method (also known as the Sanger dideoxy method after its inventor).
- Maxam–Gilbert technique depends on the relative chemical liability of different nucleotide bonds, whereas the Sanger method interrupts elongation of DNA sequences by incorporating dideoxynucleotides into the sequences.
- The chain termination method is the method more usually used because of its speed and simplicity.
Interesting Science Videos
Chemical Cleavage Method (Maxam–Gilbert Method)
- In 1976-1977, Allan Maxam and Walter Gilbert developed a DNA sequencing method based on chemical modification of DNA and subsequent cleavage at specific bases.
- The method requires radioactive labelling at one end and purification of the DNA fragment to be sequenced.
- Chemical treatment generates breaks at a small proportions of one or two of the four nucleotide based in each of four reactions (G,A+G, C, C+T).
- Thus a series of labelled fragments is generated, from the radiolabelled end to the first ‘cut’ site in each molecule.
- The fragments in the four reactions are arranged side by side in gel electrophoresis for size separation.
- To visualize the fragments, the gel is exposed to X-ray film for autoradiography, yielding a series of dark bands each corresponding to a radiolabelled DNA fragment, from which the sequence may be inferred.
Key Features
- Base-specific cleavage of DNA by certain chemicals
- Four different chemicals, one for each base
- A set of DNA fragments of different sizes
- DNA fragments contain up to 500 nucleotides
Advantages
- Purified DNA can be read directly
- Homopolymeric DNA runs are sequenced as efficiently as heterogeneous DNA sequences
- Can be used to analyze DNA protein interactions (i.e. footprinting)
- Can be used to analyze nucleic acid structure and epigenetic modifications to DNA
Disadvantages
- It requires extensive use of hazardous chemicals.
- It has a relatively complex set up / technical complexity.
- It is difficult to “scale up” and cannot be used to analyze more than 500 base pairs.
- The read length decreases from incomplete cleavage reactions.
- It is difficult to make Maxam-Gilbert sequencing based DNA kits.
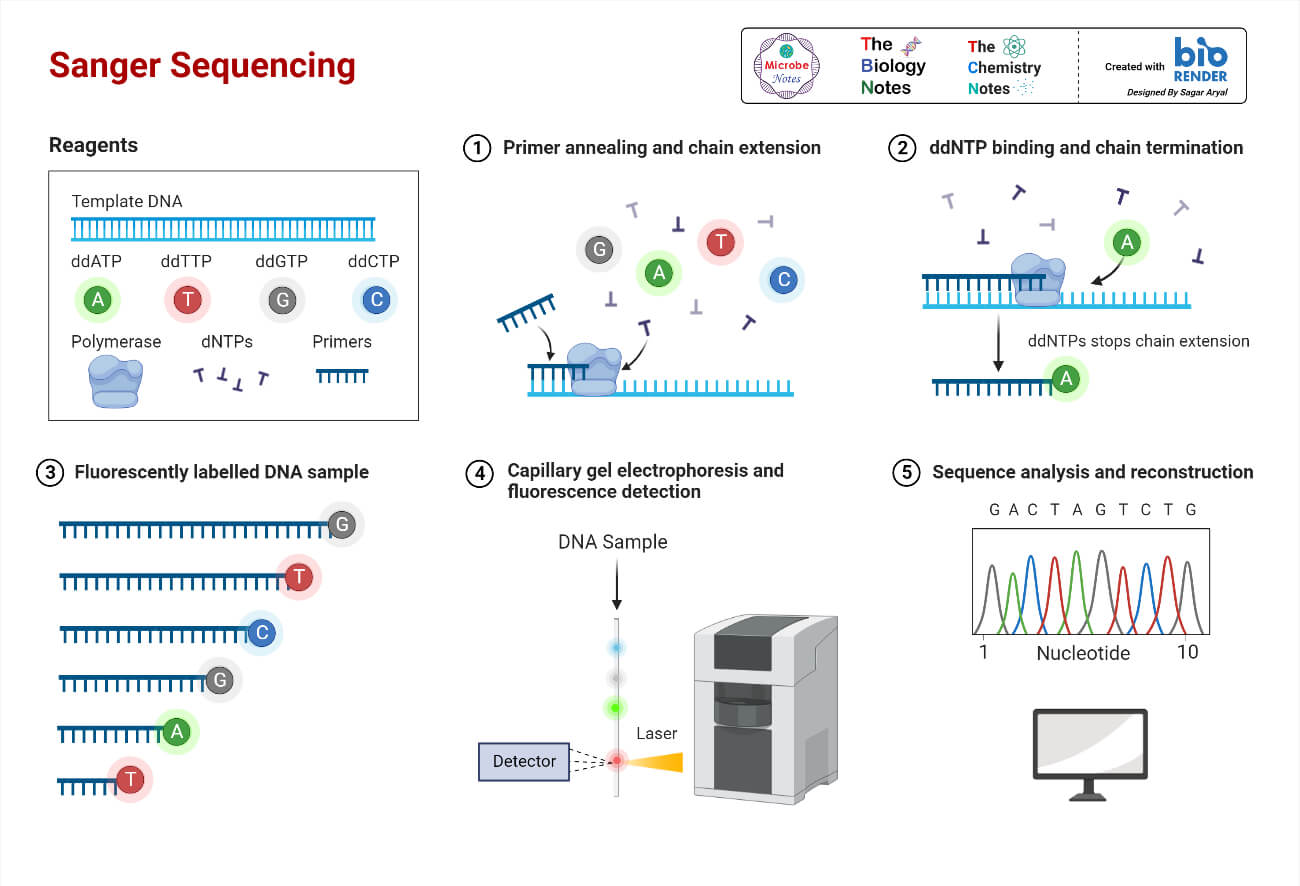
Chain Termination Method (Sanger Dideoxy Method)
- The chain terminator method is more efficient and uses fewer toxic chemicals and lower amount of radioactivity than the method of Maxam and Gilbert.
- The key principle of the Sanger method was the use of dideoxynucleotide triphosphates (ddNTPs) as DNA chain terminators.
- The chain termination method requires a single-stranded DNA template, a DNA primer,a DNA polymerase, radioactively or fluorescently labelled nucleotides,and modified nucleotides that terminate DNA strand elongation.
- The DNA sample is divided into four separate sequencing reactions, containing all four of the standard deoxynucleotides (dATP, dGTP, dCTP, dTTP) and the DNA polymerase.
- To each reaction is added only one of the four dideoxynucleotide (ddATP, ddGTP, ddCTP, ddTTP) which are the chain terminating nucleotides, lacking a 3’-OH group required for the formation of a phosphodiester bond between two nucleotides, thus terminating DNA strand extension and resulting in DNA fragments of varying length.
- The newly synthesized and labelled DNA fragments are heat denatured, and separated by size by gel electrophoresis on a denaturing polyacrylamide-urea gel with each of the four reactions run in one of the four individual lanes (lanes A, T, G, C).
- The DNA bands are then visualized by autoradiography or UV light, and the DNA sequence can be directly read off the X-ray film or gel image.
- A dark band in a lane indicates a DNA fragment that is result of chain termination after incorporation of a dideoxynucleotide (ddATP, ddGTP, ddCTP, or ddTTP).
- The relative position of the different bands among the four lanes are then used to read (from bottom to top) the DNA sequence.
- The technical variations of chain termination sequencing include tagging with nucleotides containing radioactive phosphorus for labelling, or using a primer labelled at the 5’ end with a fluorescent dye.
- Dye- primer sequencing facilitates reading in an optical system for faster and more economical analysis and automation.
Key Features
- Uses dideoxy nucleotides to terminate DNA synthesis.
- DNA synthesis reactions in four separate tubes
- Radioactive dATP is also included in all the tubes so the DNA products will be radioactive.
- Yielding a series of DNA fragments whose sizes can be measured by electrophoresis.
- The last base in each of these fragments is known.
Advantage
Chain termination methods have greatly simplified DNA sequencing.
Limitations
- Non-specific binding of the primer to the DNA, affecting accurate read-out of the DNA sequence.
- DNA secondary structures affecting the fidelity of the sequence.
Significance of DNA Sequencing
- Information obtained by DNA sequencing makes it possible to understand or alter the function of genes.
- DNA sequence analysis demonstrates regulatory regions that control gene expression and genetic “hot spots” particularly susceptible to mutation.
- Comparison of DNA sequences shows evolutionary relationships that provide a framework for definite classification of microorganisms including viruses.
- Comparison of DNA sequences facilitates identification of conserved regions, which are useful for development of specific hybridization probes to detect microorganisms including viruses in clinical samples.
- DNA sequencing has become sufficiently fast and inexpensive to allow laboratory determination of microbial sequences for identification of microbes. Sequencing of the 16S ribosomal subunit can be used to identify specific bacteria. Sequencing of viruses can be used to identify the virus and distinguish different strains.
- DNA sequencing shows gene structure that helps research workers to find out the structure of gene products.
References
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
- http://library.umac.mo/ebooks/b28050393.pdf
- http://www.aun.edu.eg/molecular_biology/Proceeding_Dec2011/DNA%20sequencing.pdf
- https://ab.inf.uni-tuebingen.de/teaching/ws09/bioinformatics-i/10-sequencing.pdf
- http://www.pathologyoutlines.com/topic/molecularpathdnaseqmaxam.html

Microbe notes is really great. It has always been there in time of need. Great and thanks for your support and service 🙏🏻
Nicely done— but the colored diagram under the Sanger method has the —gel labeled incorrectly—- more specifically— looking at the bottom of the pictured gel— the bottom nucleotides — left to right go C,T,A,G— but— should be— G,A,T,C— then the gel read through corresponds to the lanes— !
Otherwise— really nicely done!
Wow..thanks