Interesting Science Videos
What is pectin?
- Pectin is a complex heteropolysaccharide composed of linear chains of α-D-galacturonic acid or other similar sugar derivatives, commonly found in plant cell walls as cementing material.
- Pectin often remains associated with other cell wall polysaccharides like cellulose, hemicelluloses, and lignin.
- The highest concentration of pectin is found in the primary cell wall and middle lamella of plant cells with decreasing concentration towards the plasma membrane.
- Pectin is responsible for providing firmness and structure to the cell wall and also helps in intercellular adhesion and mechanical resistance of the cell.
- Most of the natural pectin is water-soluble or free; however, some forms of non-soluble or bound pectin can also be found.
- The degree of solubility of pectin depends on the length of the polymer and the presence of a methoxy group in the structure.
- Because pectin can exist as a thick gel-like structure, the commercial application of pectin is extensive.
- Pectin is one of the few biopolymers that are studied because of its high fermentable dietary fibers.
- Pectin has multiple applications because of their structural diversity and complexity.
- The term ‘pectin’ is derived from the Greek word ‘pektikos’ which means curdled or congealed.
Structure of pectin
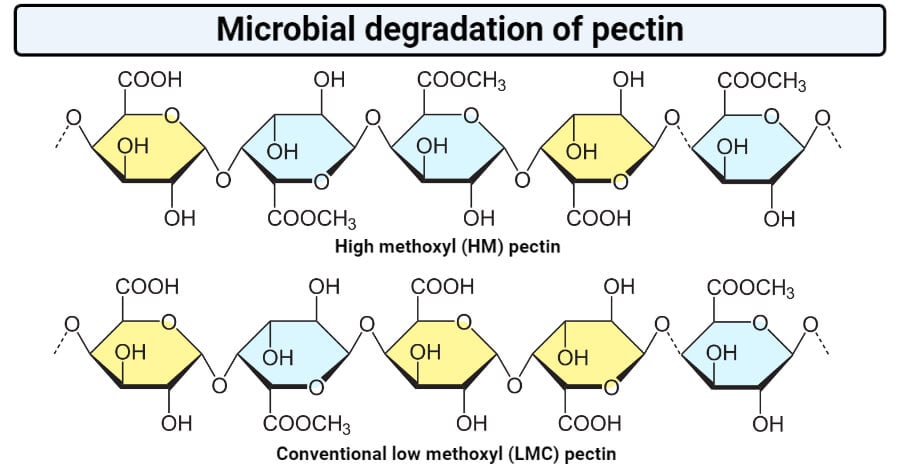
Image Source: Silvateam.
- Pectins are a family of covalently associated galacturonic acid-rich plant cell wall polysaccharides.
- About 70% of all pectin contains galacturonic acid and all pectin polysaccharides contain galacturonic acid linked at the O-1 and O-4 positions of the polymer.
- The structural elements of pectin are classified into two families; galacturonans and rhamnogalacturonan.
- Galactorunans consist of a backbone of α-(1,4)-linked D-galacturonic acid residues. The branched can either be branched or unbranched.
- The backbone in rhamnogalacturonans, however, contains diglycosyl repeating units of α-L-rhamnose-(1,4)-α-D-galacturonic acid.
- The rhamnose residues are ramified at the O-4 and O-3 positions with polymeric side chains that include arabinose and galactose residues at other positions.
- In pectin, four different types of polymeric side chains might exist; arabinans, galactans, type I arabinogalactans, and type II arabinogalactans.
- The chemical structure of pectin is extremely complicated as it contains as many as 18 different monosaccharides linked together by twenty different linkages.
- The overall structure of pectin is explained in terms of smooth and hairy regions. The smooth regions contain linear chains of homo or heteropolymer, whereas the hairy region contains simple or complex side chains.
Besides, several other monosaccharides might remain bonded by modified O-ether or O-ester linkages.
1. Galacturonans
- The galacturonans found in pectin can either be homogalacturonans or hetero galacturonans.
- Homogalacturonans form the smooth region of pectin, that is unbranched chains of α-(1,4)-linked G-galacturonic acid residues that might be methyl or acetyl-esterified.
- Homogalacturonans accounts for about 60% of all pectin found in different living beings.
- In the case of heterogalacturonanas, the homopolysaccharide chain is more or less heavily substituted at O-2 and O-3 by monomers or dimers of xylose, resulting in axylogalacturonan.
- If the polymer is substituted with complex side chains like rhamnose, it forms rhamnogalacturonans.
2. Rhamnogalacturonans
- Some of the pectins might exist as rhamnogalacturonan consisting of a long chain of alternating L-rhamnose and D-galacturonic acid residues.
- In some cases, the rhamnose residues might even be replaced by a variety of L-arabinosyl and D-galactosyl-containing side chains.
- A small number of glucuronic acid and 4-O-methyl glucuronic acid residues might be present.
- Rhamnogalacturonans account for about 20-35% of the total pectin content in nature, but the amount is as high as 75% in the soybean plant.
What are Pectinases?
- Pectinases is a group of enzymes (at least seven different enzymes) involved in the breakdown of pectin obtained from various sources.
- Due to the diverse group of pectin that is found in different living organisms, pectinases are also diverse.
- Most common and industrially important pectinases are divided into different groups on the basis of the differences in their substrate, structure, and reaction mechanism.
- Some of the common pectinolytic enzymes include pectinesterases, polygalacturonases, pectin lyases, and pectin depolymerases.
- Pectinases have important industrial applications as they are involved in the extraction and clarification of juice and maceration of plant tissues.
- Pectinases are one of the major enzymes that take in the global carbon cycle, assisting in natural waste recycling.
- Pectinases are even termed carbon recycling agents in nature as they degrade pectin substances into saturated and unsaturated galacturonans, which can then be catabolized to form either pyruvate or 3-phosphoglyceraldehyde.
- Along with these applications, pectinases are also used in degumming fiber crops, as enzyme complex for the generation of animal feed, purifying plant viruses, and in the extraction of oils.
- Some pectinases act as virulence factors as these enzymes help in the degradation of pectin found in the cell of plants.
- Pectinases might differ in structure and mechanism on the basis of their source, like the pectinase from fungi might be different from that of bacteria.
- Bacterial pectinases tend to be alkaline, whereas fungal pectinases are acidic in nature.
Microorganisms involved in pectin degradation (pectinolytic microorganisms)
Different groups of microorganisms are known to produce multiple sets of pectinolytic enzymes that aid either in the absorption of nutrients or help in the pathogenesis of microbial diseases.
1. Pectinolytic bacteria
- Bacteria have recently become a major source of pectinolytic enzymes where they produce different sets of enzymes that help in the overall degradation of pectin substrates.
- Some of the common pectinolytic bacteria include organisms like Bacillus, Pseudomonas, and Staphylococcus.
- Most of the bacterial pectinolytic activity is observed under aerobic conditions by aerobes, whereas some of the activity might be seen under anaerobic conditions.
- Some bacteria like Bacillus badius, Bacillus asahin, Bacillus psychrosaccharolyticus, and Pseudomonas aeruginosa even utilize the pectinolytic activity in their pathogenesis of different diseases.
- Common thermophilic bacteria like Geobacillus sp, Anoxybacillus sp, and Bacteroides also exhibit pectinase activity, assisting in the recycling of carbon compounds in the biosphere.
2. Pectinolytic fungi
- Fungi are the most group of microorganisms involved in the degradation of polysaccharides as a part of the natural recycling process.
- These fungi might exist in different habitats with different lifestyles.
- The most common group of fungi involved in pectin degradation are the species belonging to Ascomycetes and Deuteromycetes.
- Phanerochaete chrsosporium is one of the most studied basidiomycetes (white rot) fungi that degrade most of the complex polysaccharides like cellulose, pectin, and chitin.
- Other fungal species involved in pectin degradation include Magnaporthe oryzae, Giberella zeae, Botrytis fuckeliana, Sclerotinia sclerotiorum, Aspergillus nidulans, Trichoderma virens, Podospora anserine, Rhizopus oryzae, and Aspergillus clavatus.
- The type of enzymes produced and their mode of action might differ with the fungal species.
Enzymes involved in the degradation of pectin
Depending on the source of the enzymes, their substrates, and the reaction mechanism, pectinases are classified into different groups;
1. Polygalacturonase
- Polygalacturonase is a group of enzymes that hydrolyze O-glycosyl bonds in the homogalacturonan to form monomeric units.
- These enzymes act on the 1,4-α-D-galactosyluronic linkages between the galacturonic residues.
- Most of the polygalacturonases are endo-enzymes that act on the linkages randomly to depolymerize the chain or reduce the length of the polymer.
- The natural substrate of endo-polygalacturonase is homogalacturonan; however, other compounds like oligogalacturonides might also act as a substrate depending on the nature of the substrate.
- A class of exo-polygalacturonase is also known where they break down the polygalacturonates into di- and mono-galacturonates.
- The activity of the enzyme is determined by the measurement of reducing sugars formed as a result of hydrolysis or by the viscous reduction method.
2. Pectinesterase
- Pectinesterases are a group of enzymes that catalyzes the hydrolysis of methylated carboxylic ester in pectin to form pectic acid and methanol.
- The natural substrate of pectinesterase is pectin; however, other compounds like methyl pectate and methylated oligogalacturonides also work as substrate.
- The activity of pectinesterase is enhanced or induced by (NH4)2SO4, Mg2+, and NaCl. It is inhibited by the presence of Cu2+ and Hg2+.
- Most of the well-studied pectinesterase is produced from plants; however, recently pectinesterase of bacterial and fungal origin have also been discovered.
- Most pectinases are specific towards esterified pectic substances and thud, might not show any activity towards pectates.
3. Pectin lyases
- Pectin lyases degrade pectin substances in a random fashion, yielding a 4:5 ratio of unsaturated oligomethylgalacturonates.
- These enzymes cleave glycosidic linkages, preferentially on polygalacturonic acid through transelimination reaction.
- Pectin lyases have an absolute requirement of Ca2+ ions and thus, are inhibited by chelating agents like EDTA.
- Exo-pectin lyases catalyze the cleavage of the substrate from the non-reducing end of the polymer.
Factors affecting pectin degradation
Pectin degradation in nature and on artificial growth media is affected by different factors, some of which are:
1. Moisture content
- Based on studies done on pectin degradation, it has been observed that the rate of chitin degradation is rapid in the presence of free water and complete saturation.
- The change in water concentration or moisture content has minimal effect on the rate of pectin degradation.
- Nevertheless, the rate is impaired if the amount of water increases to the point that causes impairing of aeration due to logging.
2. Aeration
- Most of the pectinolytic microorganisms are aerobic or facultative aerobic. As a result, the rate of pectin utilization or degradation is enhanced in the increased concentration of O2.
- Some amount of degradation, however, can be achieved in a low concentration of CO2 as it allows facultative aerobes and anaerobes to remain active.
- Pure oxygen environment (100% O2) might be toxic in some cases, especially when readily energy sources are available.
3. Added glucose
- The addition of glucose in the media or soil hampers the rate of pectin degradation as the organisms utilize the readily available source of energy rather than pectin as their source of nutrients.
- Glucose is a ready energy source which is easy to metabolize. This, in turn, causes a delay or decreased pectin degradation.
- In the absence of glucose or such similar sources, however, pectin degradation is enhanced as pectin is comparatively less complex when compared to other carbohydrate sources like lignin.
4. Organic matter
- The presence of plant fibers rich in pectin also supports the rate of pectin degradation.
- Organic matter rich in nutrients and minerals for the microorganisms helps in the formation of biomolecules like proteins and enzymes.
- The increase in organic matter increases the substrate concentration. The increased substrate concentration might decrease the degradation rate at first as the organisms utilize sources like glucose and cellulose.
- As these sources are degraded, pectin becomes the next source of nutrition, which then increases its’ degradation.
Process (Simple Steps) of pectin degradation
The microbial hydrolysis or degradation of pectin in nature occurs in the form of the following steps;
1. Deesterification
- The first enzyme acting on pectin substances is pectin esterases or pectin methyl esterases.
- These enzymes catalyze the deesrerification of the methoxy group of pectin, resulting in pectic acid and methanol.
- Esterase enzymes act prior to polygalacturonates and pectate lyases as they need non-esterified substrates.
- Pectin esterases prefer a methyl ester group of galacturonate units next to a non-esterified galacturonate unit.
- Esterases like pectin acetyl esterases hydrolyze the acetyl ester of pectin, resulting in pectic acid and acetate.
2. Hydrolytic cleavage
- The most important step of pectin degradation is the hydrolytic cleavage of the α-1,4-glycosidic linkages that exist in the backbone of the pectin substrates.
- For this, different enzymes are produced by different microorganisms, which act on a different group of pectin substrate.
- Polymethylgalacturonases and polygalacturonases act on the α-1,4-glycosidic linkages on highly esterified pectin, resulting in 6-methyl-D-galacturonate and D-galacturonate respectively.
- Both of these enzymes can act as either endo or exoenzymes cleaving the pectin backbone either randomly or through the reducing ends.
- Another group of hydrolytic enzymes is pectate lyases that act on the glycosidic linkages of polygalacturonic acid, forming unsaturated products through transelimination reaction.
Mechanisms of microbial degradation of pectin
- The mechanism of microbial degradation of pectin differs with the type of enzyme involved in the process.
- The following are some mechanism of action of enzymes involved in pectin degradation:
1. Mechanism of de-esterification by pectin esterases and pectin methyl esterases
Pectin esterases act on the pectin substrates by one of the three mechanisms;
- The single-chain mechanism where the enzyme acts on all substrate side on the polymeric chain.
- The multiple-chain mechanism involving the catalysis of just one reaction which then dissociates the substrate.
- A multiple-attack mechanism where the enzyme catalyzes multiple reactions before the enzyme-substrate complex dissociates.
- Bacterial polyesterases produce products with adjacent regions of galacturonic acids via both single-chain and multiple-attack mechanism.
- Fungal esterases, however, attack randomly by a multiple-chain mechanism.
- During the random attack, de-esterification causes the release of protons which promotes the action of endopolygalacturonases.
Example
- The mechanism of de-esterification of galacturonan macromolecules in Bacteroides occurs by a multi-attack mechanism, which is then followed by the decomposition of the oligomers to release the end products.
2. Mechanism of hydrolytic cleavage in polygalacturonases and pectin lyases
- The process of hydrolytic cleavage of α-1,4-glycosidic bonds in pectin begins with the positioning of the active site amino acids on the susceptible glycosidic bonds.
- The motifs on the active sites interact with the substrate on either side of the designated bond through multiple hydrogen bonds.
- The hydrogen bonds create sufficient strain and distortion on the susceptible glycosidic bond.
- The distortion is followed by proton transfer between the amino acids of the active site and the glycosidic bond.
- This causes the cleavage of glycosidic bonds with the release of the first end product with subsequent formation of a covalent bond between the substrate and the catalytic site nucleophile.
- Another active site residue on the enzyme then places a water molecule for a nucleophilic attack on the substrate. The nucleophilic attack results in the formation of the second end product, along with the restoration of the active site of the enzyme.
Example
- Hydrolytic cleavage by a set of polygalacturonases in Rhizopus oryzae involves about 18 polygalacturonases and one β-galactosidase. These enzymes cleave the α-1,4-glycosidic linkages by the above-mentioned mechanism by endo and exoenzymes.
References
- Ropartz D., Ralet, MC. (2020) Pectin Structure. In: Kontogiorgos V. (eds) Pectin: Technological and Physiological Properties. Springer, Cham. https://doi.org/10.1007/978-3-030-53421-9_2
- Onumpai C, Kolida S, Bonnin E, Rastall RA. Microbial utilization and selectivity of pectin fractions with various structures. Appl Environ Microbiol. 2011;77(16):5747-5754. doi:10.1128/AEM.00179-11
- Pacheco MT, Villamiel M, Moreno R, Moreno FJ. Structural and Rheological Properties of Pectins Extracted from Industrial Sugar Beet By-Products. Molecules. 2019;24(3):392. Published 2019 Jan 22. doi:10.3390/molecules24030392
- Mellinas C, Ramos M, Jiménez A, Garrigós MC. Recent Trends in the Use of Pectin from Agro-Waste Residues as a Natural-Based Biopolymer for Food Packaging Applications. Materials (Basel). 2020;13(3):673. Published 2020 Feb 3. doi:10.3390/ma13030673
- Minzanova ST, Mironov VF, Arkhipova DM, et al. Biological Activity and Pharmacological Application of Pectic Polysaccharides: A Review. Polymers (Basel). 2018;10(12):1407. Published 2018 Dec 19. doi:10.3390/polym10121407
- Lara-Espinoza C, Carvajal-Millán E, Balandrán-Quintana R, López-Franco Y, Rascón-Chu A. Pectin and Pectin-Based Composite Materials: Beyond Food Texture. Molecules. 2018;23(4):942. Published 2018 Apr 18. doi:10.3390/molecules23040942
- Thakur BR, Singh RK, Handa AK. Chemistry and uses of pectin–a review. Crit Rev Food Sci Nutr. 1997 Feb;37(1):47-73. DOI: 10.1080/10408399709527767. PMID: 9067088.
- MOHNEN, D. (2008). Pectin structure and biosynthesis. Current Opinion in Plant Biology, 11(3), 266–277.doi:10.1016/j.pbi.2008.03.006
- Abbott DW, Boraston AB. Structural biology of pectin degradation by Enterobacteriaceae. Microbiol Mol Biol Rev. 2008;72(2):301-316. doi:10.1128/MMBR.00038-07
- Favela-Torres E., Aguilar C., Contreras-Esquivel J., Viniegra-González G. (2006) Pectinases. In: Pandey A., Webb C., Soccol C.R., Larroche C. (eds) Enzyme Technology. Springer, New York, NY. https://doi.org/10.1007/978-0-387-35141-4_14
- Gummadi S.N., Manoj N., Kumar D.S. (2007) Structural and Biochemical Properties of Pectinases. In: Polaina J., MacCabe A.P. (eds) Industrial Enzymes. Springer, Dordrecht. https://doi.org/10.1007/1-4020-5377-0_7
- Pedrolli, Danielle & Monteiro, Ana & Gomes, Eleni & Carmona, Eleonora. (, 2009). Pectin and Pectinases: Production, Characterization and Industrial Application of Microbial Pectinolytic Enzymes. The Open Biotechnology Journal. 3. 9-18. 10.2174/1874070700903010009.
- Benoit, Isabelle & Coutinho, Pedro & Schols, H.A. & Gerlach, Jan & Henrissat, Bernard & Vries, R.P.. (2012). Degradation of different pectins by fungi: Correlations and contrasts between the pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC genomics. 13. 321. 10.1186/1471-2164-13-321.
- Aaisha, G.A., and Barate, D.L. 2016. Isolation and Identification of Pectinolytic Bacteria from Soil Samples of Akola Region, India. Int.J.Curr.Microbiol.App.Sci. 5(1): 514-521 http://dx.doi.org/10.20546/ijcmas.2016.501.051
- Yadav, Sangeeta & Yadav, Pramod & Yadav, Dinesh & Yadav, Kapil. (2009). Pectin lyase: A review. Process Biochemistry. 44. 1-10. 10.1016/j.procbio.2008.09.012.
- Kohli P, Kalia M, Gupta R (2015) Pectin Methylesterases: A Review. J Bioprocess Biotech 5: 227 doi:10.4172/2155-9821.1000227
- Palanivelu P (2006). Polygalacturonases: Active site analysis and mechanism of action. Indian Journal of Biotechnology. Vol 5, April 2006, 148-162.
- Suberkropp K. (2005) Pectin-Degrading Enzymes: Polygalacturonase and Pectin Lyase. In: Graça M.A., Bärlocher F., Gessner M.O. (eds) Methods to Study Litter Decomposition. Springer, Dordrecht. https://doi.org/10.1007/1-4020-3466-0_36.
Sources
- 1% – https://www.sciencedirect.com/topics/chemistry/deesterification
- 1% – https://pdfs.semanticscholar.org/39e4/291048774177355ecbfa0f60fa62f389e816.pdf
- 1% – http://www.esalq.usp.br/lepse/imgs/conteudo_thumb/Pectin-structure-and-biosynthesis.pdf
- <1% – https://www.sciencedirect.com/topics/neuroscience/pectin
- <1% – https://www.sciencedirect.com/topics/medicine-and-dentistry/bacteroidetes
- <1% – https://www.sciencedirect.com/topics/chemistry/rhamnogalacturonan
- <1% – https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/pectin
- <1% – https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/pectinase
- <1% – https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/pectinesterase
- <1% – https://www.sciencedirect.com/science/article/pii/S0734975008001092
- <1% – https://www.sciencedirect.com/science/article/pii/S0734975011000681
- <1% – https://www.sciencedirect.com/science/article/pii/S0141391018302854
- <1% – https://www.researchgate.net/publication/315676636_Water_Determination
- <1% – https://www.researchgate.net/publication/240512252_Postharvest_softening_of_apple_Malus_domestica_fruit_A_review
- <1% – https://www.researchgate.net/publication/226654996_Structural_and_Biochemical_Properties_of_Pectinases
- <1% – https://www.researchgate.net/publication/12374470_Analysis_of_different_de-esterification_mechanisms_for_pectin_by_enzymatic_fingerprinting_using_endopectin_lyase_and_endopolygalacturonase_II_from_A_Niger
- <1% – https://www.researchgate.net/profile/RP_Vries/publication/229425157_Degradation_of_different_pectins_by_fungi_Correlations_and_contrasts_between_the_pectinolytic_enzyme_sets_identified_in_genomes_and_the_growth_on_pectins_of_different_origin/links/00463528d373a0a54b000000/Degradation-of-different-pectins-by-fungi-Correlations-and-contrasts-between-the-pectinolytic-enzyme-sets-identified-in-genomes-and-the-growth-on-pectins-of-different-origin.pdf
- <1% – https://www.ijcmas.com/vol-5-1/G.A.%20Aaisha%20and%20D.L.%20Barate.pdf
- <1% – https://www.hindawi.com/journals/ijmicro/2009/631942/
- <1% – https://www.chempap.org/file_access.php?file=382a223.pdf
- <1% – https://sfamjournals.onlinelibrary.wiley.com/doi/abs/10.1046/j.1472-765X.2002.01159.x
- <1% – https://quizlet.com/80828639/zoology-final-part-1-flash-cards/
- <1% – https://pediaa.com/what-is-the-active-site-of-an-enzyme/
- <1% – https://openagricola.nal.usda.gov/catalog/CAIN709030374
- <1% – https://medical-dictionary.thefreedictionary.com/pectinesterase
- <1% – https://link.springer.com/article/10.1007/s11224-009-9442-z
- <1% – https://journals.sagepub.com/doi/pdf/10.1177/000456328602300602
- <1% – https://infinitabiotech.com/blog/uses-of-pectin-methylesterase/
- <1% – https://en.wikipedia.org/wiki/Polygalacturonase
- <1% – https://drrathresearch.org/images/attachments/education/Phytobiology/Pectin.pdf
- <1% – https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-13-321
- <1% – http://www.plantphysiol.org/content/153/2/384
- <1% – http://www.drrathresearch.org/images/attachments/education/Phytobiology/Pectin.pdf
- <1% – http://people.uleth.ca/~steven.mosimann/bchm3100/Bchm3100_L9.pdf
