Staphylococcus hominis is a Gram-positive coagulase-negative member of Staphylococci that exists as a commensal on the human body, especially in the areas with apocrine glands, axillae, and the pubic region.
- Like most other coagulase-negative Staphylococci, S. hominis is also known to cause various nosocomial or hospital-acquired infections and may occasionally cause infection in patients with abnormally weak immune systems
- The term ‘hominis’ is derived from the Latin term ‘hominis’ which means humans, and it thus is named for the host on whose skin this species is commonly found.
- Like all Staphylococci, S. hominis is also clustering Gram-positive cocci, nonmotile, non-spore-forming, and facultatively anaerobic.
- It has been designated as a potential pathogen, but so far the exact pathogenic mechanisms of this bacterium have not been determined.
- S. hominis has been further divided into two subspecies; S. hominis subsp. hominis and novobiosepticus.
- S. hominis subsp. hominis was first isolated by Kloos and Schleifer in 1975 whereas S. hominis subsp. novobiosepticus was first isolated in 1996 by Kloos.
- Strains of S. hominis are known to colonize the skin of a person for relatively short periods of time, usually several weeks to several months, compared to many of the strains of the predominant species Staphylococcus epidermidis that persist for one to several years.
- S. hominis has been of importance lately as it has been found to be a multi-drug resistance strain.
Image Source: Vetbact, created with biorender.com.
Interesting Science Videos
Classification of Staphylococcus hominis
The species of Staphylococci are classified into different species primarily on the basis of DNA–DNA hybridization. Besides, other characteristics like fatty acids composition and G+C content are also observed. The subspecies of S. hominis are further divided on the basis of their activity against novobiocin and their habitat. S. hominis subsp. hominis consists of strains that are susceptible towards novobiocin and are primarily found on the skin surface, whereas S. hominis subsp. novobiosepticus is resistant to novobiocin and can be isolated from blood.
| Domain: | Bacteria |
| Phylum: | Firmicutes |
| Class: | Bacilli |
| Order: | Bacillales |
| Family: | Staphylococcaceae |
| Genus: | Staphylococcus |
| Species: | S. hominis |
| Subspecies: | S. hominis subsp. hominis |
| Subspecies: | S. hominis subsp. novobiosepticus |
Habitat of Staphylococcus hominis
- Humans are the primary host for both of the subspecies of S. hominis as these are mostly found as commensal organisms on the skin surface.
- While other coagulase-negative Staphylococci like S. epidermidis colonize the upper part of the body, S. hominis is mostly found on the lower part of the body like the perineal and groin areas.
- It is found in a large number in areas with numerous apocrine glands that retain some amount of moisture.
- In a recent study, it was found that S. hominis account for about 22% of all Staphylococci species found on the human skin.
- Besides, these are also found on the scalp of preadolescent children along with other species of Staphylococci like S. capitis.
- S. hominis, unlike other Staphylococci species like S. lugdunensis, is found in virtually all parts of the body in different numbers. The numbers also change over a few weeks as they tend to colonize certain areas for a shorter period of time.
Morphology of Staphylococcus hominis
- Both the species of S. hominis are Gram-positive, nonmotile, non-spore-forming cocci with an average size of 1.0–1.5 μm in diameter.
- The arrangement of the cells is characteristic of all Staphylococci species where the organisms occur singly or form tetrads and smaller numbers of pairs. This arrangement is due to the property of the organism to divide in more than one plane to form irregular grapelike clusters.
- These are facultatively anaerobic but show weak and delayed growth under anaerobic conditions.
- Unlike coagulase-positive Staphylococci like S. aureus, S. hominis doesn’t have a capsule surrounding the cell wall.
- The cell wall, however, is composed of characteristic peptidoglycan and teichoic acid that provides shape and protection to the cell.
- The cell membrane is made up of a lipid-protein bilayer composed of peptidoglycan and other proteins.
- Like all other coagulase-negative Staphylococci, S. hominis also has fewer cell wall adhesions and cell-wall associated proteins.
Cultural Characteristics of Staphylococcus hominis
The selective media for most Staphylococci species includes media like P agar, Mannitol Salt Agar, Baird-Parker agar, and liquid medium like thioglycollate medium. Even though the organism is facultatively anaerobic, it shows weak or delayed growth under anaerobic conditions. The optimum temperature for growth is 37°C, but some growth is seen from 20°C to 45°C. Sufficient growth can be seen at 10% NaCl with decreased growth at 15%.
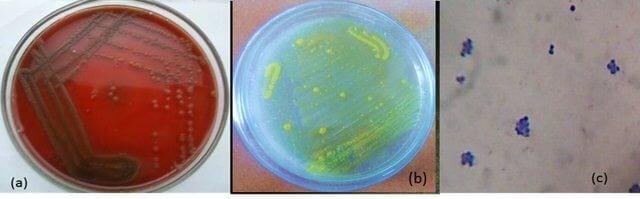
Figure: Staphylococcus hominis MSD1 on (a) Blood agar (b)Mannitol salt agar (c) Gram-positive cocci in clusters under 100X light microscopy. Image Source: Subathra Devi C.
The following is the colony morphology observed on different media:
1. Nutrient Agar (NA)
- Circular, cream-colored to white colonies of S. hominis are observed on NA. The colonies are mostly 1mm in diameter with an entire margin.
- The colonies have raised elevation and a dense center with transparent borders.
2. Mannitol Salt Agar (MSA)
- Small pink to red colonies are formed on MSA. The media remains red as the bacterium cannot ferment mannitol.
- The colonies are 1-2 mm in diameter with an entire margin.
3. P agar
- S. hominis subsp. hominis: Colorless to cream to yellow-orange colonies of diameter 3-5 mm are seen on P agar. The colonies are smooth, opaque, raised to umbonate, and butyrous, with entire margins.
- S. hominis subsp. novobiosepticus: Grayish-white, convex to umbonate, butyrous, and opaque colonies, with entire margins. Colonies of the size 4–6 mm in diameter after incubation at 34–35°C for three days.
Biochemical characteristics of Staphylococcus hominis
The biochemical characteristics of S. hominis can be tabulated as follows:
| S.N | Biochemical Characteristics | S. hominis |
| 1. | Capsule | No capsule |
| 2. | Shape | Cocci |
| 3. | Catalase | Positive (+) |
| 4. | Oxidase | Negative (-) |
| 5. | Citrate | Negative (-) |
| 6. | Methyl Red (MR) | Negative (-) |
| 7. | Voges Proskauer (VR) | Negative (-) |
| 8. | Urease | Positive (+) |
| 9. | Coagulase | Negative (-) |
| 10. | DNase | Negative (-) |
| 11. | Clumping factor | Negative (-) |
| 12. | Gas | Positive (+) |
| 11. | H2S | Positive (+) |
| 12. | Hemolysis | Negative (-) |
| 13. | Motility | Negative (-) |
| 14. | Nitrate Reduction | Positive (+) |
| 15. | Gelatin Hydrolysis | Negative (-) |
| 16. | Pigment Production | Variable |
| 17. | Novobiocin resistance | Susceptible (S. hominis subsp. hominis), Resistant (S. hominis subsp. novobiosepticus) |
| 18. | Bile esculin test | Negative (-) |
Fermentation
| S.N | Substrate | S. hominis |
| 1. | Mannitol | Negative (-) |
| 2. | Glucose | Positive (+) May produce only d-lactate or both l- and d-lactate from glucose anaerobically. |
| 3. | Fructose | Positive (+) |
| 4. | Galactose | Positive (+) |
| 5. | Lactose | Positive (+) |
| 6. | Maltose | Positive (+) |
| 7. | Mannose | Negative (+) |
| 8. | Raffinose | Negative (-) |
| 9. | Ribose | Negative (-) |
| 10. | Sucrose | Positive (+) |
| 11. | Starch | Negative (-) |
| 12. | Trehalose | Negative (+) |
| 13. | Xylose | Negative (-) |
| 14. | Salicin | Positive (-) |
| 15. | Glycerol | Positive (+) |
| 16. | Dulcitol | Negative (-) |
| 17. | Cellobiose | Negative (-) |
| 18. | Rhamnose | Negative (-) |
| 19. | Arabinose | Negative (-) |
| 20. | Inulin | Negative (-) |
| 21. | Sorbitol | Negative (-) |
| 22. | Pyruvate | Negative (-) |
Enzymatic Reactions
| S.N | Enzymes | S. hominis |
| 1. | Hyaluronidase | Variable |
| 2. | Acetoin | Positive (+) |
| 3. | Alkaline Phosphatase | Negative (-) |
| 4. | Ornithine Decarboxylase | Negative (-) |
| 5. | Pyrrolidonyl aminopeptidase | Positive (+) |
| 6. | β-galactosidase | Negative (-) |
S. hominis subsp. novobiosepticus can be differentiated from S. hominis subsp. hominis by novobiocin resistance, inability to utilize arginine, and inability to produce acid aerobically from D-trehalose and N-acetylglucosamine.
Virulence factors of Staphylococcus hominis
The exact mechanism of infections caused by S. hominis is yet not known, but it has been seen that are several factors present in the species assist in the process of infection. These virulence factors have also been a topic of interest for many research works as the organism is increasingly becoming more resistant to various antibiotics like aminoglycosides.
1. Adhesins
- The colonization of the surface is the first step towards the pathogenesis of infections caused by S. hominis.
- In the case of Staphylococcus species, adhesion to host tissue is achieved by a large family of surface proteins that bind with varying degrees of specificity to host matrix proteins, such as fibronectin, fibrinogen, vitronectin, laminin, and von Willebrand factor present on the host cell.
- This attachment followed by colonization is brought about by various proteins and cell-wall associated proteins that allow the binding of the bacteria to the cell surface.
- One of the most important binding proteins is the fibrinogen-binding protein found in most of the coagulase-negative staphylococci.
- Staphylococci surface protein (Ssp) and autolysin protein (Aas) are two cell-wall associated proteins that have the ability to bind with the fibrinogen present on the host cell surface.
- Strains of S. hominis have an excellent ability to bind to the HeLa cells in patients that have undergone chemotherapy. The exact mechanism of the binding is not yet known.
2. Invasion of epithelial cells
- Once the bacteria bind to the surface of the host cell, it then has the ability to invade the cells by release an extracellular protein that has cytotoxic activity.
- The range of toxicity might differ between different strains, but it is known to affect both the epithelial cells and the HeLa cells.
- The ability of the organism to cause invasion of epithelial cells is considered the primary mechanism for the entry of the bacteria into the bloodstream, causing sepsis and shock syndromes.
3. Genes providing antibiotic resistance
- MecA gene found in various bacteria is considered a major gene that provides antibiotic resistance to the bacteria against various groups of antibiotics.
- The mecA gene encodes a penicillin-binding protein, and as a result of mecA expression, beta-lactam antibiotics are not effective against such antibiotics.
- MecA gene has been recently found in the genome of S. hominis, which indicates the ability of the organism to cause infections similar to other MRSA.
- Besides, other genes like ant(4′)–Ia, aac(6′)/aph(2″) and aph(3′)–IIIa have also been seen in S. hominis which is probably the reason for the resistance of the organism against aminoglycosides.
4. Biofilm
- Biofilm formation is an important virulence factor for most coagulase-negative Staphylococci that cause infections related to medical device implants.
- A biofilm is a layer composed of bacteria living in an aggregated structure as cellular clusters or microcolonies along with extracellular matrices either release by the organism or derived from the environment.
- The biofilm is encapsulated in a matrix composed of an extracellular polymeric substance and is often separated by open water channels. These channels act as a circulatory system to deliver nutrients and remove metabolic waste products in and out of the biofilms.
- The biofilm allows bacteria to adhere to inert materials and also results in increased antibiotic resistance.
- Infections related to catheters and artificial valves have also been seen in the case of S. hominis, which are mostly caused due to the ability of the organism to form biofilms.
- Biofilms provide protection to the bacterial species against both the immune cells as well as the molecules of the antimicrobial drugs.
- Besides, it also helps the bacteria to adjust to the changing environmental factors.
Pathogenesis of Staphylococcus hominis
Infections associated with S. hominis are mostly nosocomial or hospital-acquired. These infections usually occur in patients that are immunocompromised or have recently undergone chemotherapy. In the case of cancer patients, the target cells of S. hominis is the HeLa cells as it has the ability to colonize and invade such cells. The exact pathogenesis of the infections cause by S. hominis is not yet fully understood; however, it is known that the genome of the organism carries several gene sequences that aid the process of infection by this bacterium.
1. Attachment/ Adhesion/ Colonization
- As a commensal, S. hominis is equipped with different surface proteins and molecules that aids in the process of attachment and colonization.
- One of the most important factors that support the attachment of the bacteria to epithelial cells is the Staphylococci surface protein (Ssp).
- Besides, there is an icaADBC-encoded polysaccharide intercellular adhesin (PIA) that further supports this attachment.
- The attachment of the bacteria to the cell surface enables the bacteria to colonize the surface and cause the invasion of the cells.
- Attachment is also the first step during the biofilm formation, which is further enhanced by various proteinaceous products that help in cell aggregation and binding with each other.
2. Epithelial cell invasion
- Invasion of epithelial cells and HeLa cells is another mechanism of infection employed by S. hominis.
- It has been studied that the organism is capable of releasing different extracellular toxins that have cytopathic effects on the epithelial cells and the HeLa cells.
- The exact composition of the toxins is not yet known, but these might be similar to the cytotoxins released by S. aureus.
3. Resistance against antibiotics
- S. hominis is capable of maintaining infections as it has various mechanisms that provide protection against different groups of antibiotics.
- MecA gene present in S. hominis is known to code for proteins that bind to penicillin or other such antimicrobial agents.
- This prevents the action of penicillin which provides resistance against such antibiotics.
- Besides, another group of genes consisting of ant(4′)-Ia gene are responsible for the resistance against aminoglycosides.
Clinical Manifestations of Staphylococcus hominis
- S. hominis is a commensal bacterium, mostly present as a part of the normal flora of the skin in humans; however, it has been recently associated with nosocomial infections, sometimes even resulting in sepsis and bloodstream infections.
- Most of the infections are related to medical device implants like valves and catheters, whereas others usually result after surgical operations.
- Endocarditis is another infection associated with S. hominis that is associated with artificial valves.
- S. hominis subsp. novobiosepticus is found to cause sepsis in neonates present in intensive care units. It has also been observed in cancer patients after chemotherapy.
- S. hominis infections are seldom lethal, but they significantly contribute to morbidity and health care costs.
- But because of S. hominis subsp. novobiosepticus is a multi-drug resistant species, it might cause severe results in immunocompromised patients.
Lab diagnosis of Staphylococcus hominis
As with most bacterial infections, the collection of clinical specimens is the first step of laboratory diagnosis. In the case of S. hominis, clinical specimens like the scabs, joint aspirates, and pus aspirated from deep sites are to be collected. Diagnosis of disease in the case of S. hominis infections are mostly related to the identification of the organism.
1. Molecular identification and biochemical characteristics
- Direct microscopic examination of these specimens may provide a rapid, presumptive report of gram-positive cocci resembling staphylococci.
- Direct observation is followed by isolation of the organism from primary clinical specimens on selective culture media like blood agar supplemented with 5 percent sheep blood, following an incubation period of 18–24 hrs at 35–37°C.
- Depending on the microscopic examination and cultural characteristics, species identification can be made.
- In order to determine the subspecies, an antimicrobial susceptibility test can be performed.
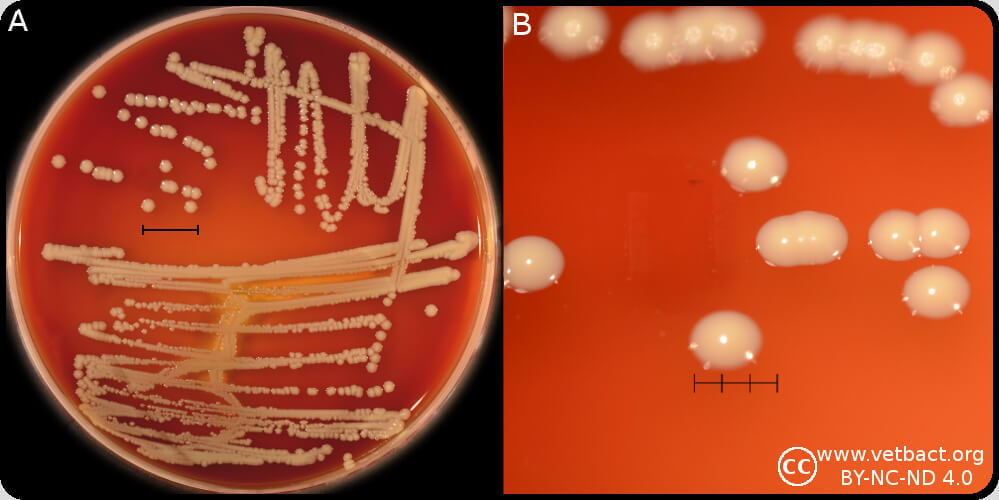
Figure: Colonies of Staphylococcus hominis, cultivated aerobically on bovine blood agar at 37°C during 24 h. The lengths of the scale bars in A and B are equivalent to 10 and 3 mm, respectively. Image Source: Vetbact.
2. Rapid identification kits
- Many clinical laboratories have started to employ different commercial identification kits or automated instruments that allow rapid determination of bacterial species.
- In the case of S. hominis, microbial cellular fatty acid compositions are used for the identification.
- Some of the common automated systems for the identification of S. hominis include MicroScan Conventional Pos ID, Rapid Pos ID, and BBL Crystal Gram-Pos ID.
3. Molecular diagnosis
- A molecular diagnosis is now considered the basis for the identification as it can provide easy and detailed identification of the species based on their nucleotide sequences.
- Real-time PCR and high-throughput DNA sequencing systems are the major molecular techniques used for the analysis of the nucleic acid sequences and identification of the species and subspecies.
- Besides, ribotyping is also a common practice that studies the rRNA by restriction fragment length polymorphism and allows molecular differentiation of S. hominis strains.
Treatment of Staphylococcus hominis
- Treatment of infections caused by S. hominis is often limited because of its resistance to different antimicrobial agents.
- Methicillin-resistant isolates and those resistant to other antimicrobials are particularly important because they have limited therapeutic options.
- Traditional antibiotic treatment protocols based on the standard in vitro susceptibility tests mostly designed for planktonic bacteria might not be applicable to eradicate biofilm-producing S. hominis infections.
- Thus, glycopeptides are usually the treatment of choice for infections caused by S. hominis.
- Besides, other forms of treatments involving the hyperimmune serum from human donors or humanized monoclonal antibodies directed towards the surface components are also being studied.
Prevention of Staphylococcus hominis
Because S. hominis is a multi-drug resistant species and is capable of forming elaborate biofilms, it is necessary to employ different resistant strategies to avoid such infections. The following are some preventive strategies that can be followed to avoid such infections:
- Coating of biomaterials or use in exit-site dressings can be employed to prevent medical devices related to infections.
- Regular cleaning and dressing wounds might also work to prevent Staph infections to a certain extent.
- The use of aseptic techniques to prevent bacterial contamination from the insertion site and catheter hubs during insertions can also be applied.
References
- Topley WWC (2007). Topley and Wison’s Microbiology and Microbial Interactions; Bacteriology, 2 Vol. Tenth Edition. John Wiley and Sons Ltd.
- Bergey, D. H., Whitman, W. B., De, V. P., Garrity, G. M., & Jones, D. (2009). Bergey’s manual of systematic bacteriology: Vol. 3. New York: Springer.
- Szczuka, E., Krzymińska, S., Bogucka, N., & Kaznowski, A. (2018). Multifactorial mechanisms of the pathogenesis of methicillin-resistant Staphylococcus hominis isolated from bloodstream infections. Antonie van Leeuwenhoek, 111(7), 1259–1265. https://doi.org/10.1007/s10482-017-1007-3Ahmed, N. H., Baruah, F. K., & Grover, R. K. (2017). Staphylococcus hominis subsp. novobiosepticus, an emerging multidrug-resistant bacterium, as a causative agent of septicaemia in cancer patients. The Indian journal of medical research, 146(3), 420–425.
- Pereira, E.M., de Mattos, C.S., dos Santos, O.C. et al. Staphylococcus hominis subspecies can be identified by SDS-PAGE or MALDI-TOF MS profiles. Sci Rep 9, 11736 (2019). https://doi.org/10.1038/s41598-019-48248-4
- Saiping Jiang, Beiwen Zheng, Wenchao Ding, Longxian Lv, Jinru Ji, Hua Zhang, Yonghong Xiao, Lanjuan Li. Whole-Genome Sequence of Staphylococcus hominis, an Opportunistic Pathogen. Journal of Bacteriology Aug 2012, 194 (17) 4761-4762; DOI: 10.1128/JB.00991-12
- Fernando Chaves, Mónica García-Álvarez, Francisca Sanz, Concepción Alba, Joaquín R. Otero. Nosocomial Spread of a Staphylococcus hominis subsp. novobiosepticus Strain Causing Sepsis in a Neonatal Intensive Care Unit. Journal of Clinical Microbiology Sep 2005, 43 (9) 4877-4879; DOI: 10.1128/JCM.43.9.4877-4879.2005
- Roy Priyamvada, Ahmed Nishat Hussain, Biswal Indu, Grover Rajesh Kumar. Multidrug-resistant Staphylococcus hominis subsp. novobiosepticus causing septicemia in patients with malignancy. Indian Journal of Pathology and Microbiology. 2014. 57;2.
- Kloos, W. E.; George, C. G.; Olgiate, J. S.; Van Pelt, L.; McKinnon, M. L.; Zimmer, B. L.; Muller, E.; Weinstein, M. P.; Mirrett, S. (July 1998). “Staphylococcus hominis subsp. novobiosepticus subsp. nov., a novel trehalose- and N-acetyl-D-glucosamine-negative, novobiocin- and multiple-antibiotic-resistant subspecies isolated from human blood cultures”. International Journal of Systematic Bacteriology. 48 (3): 799–812. doi:10.1099/00207713-48-3-799.
- Otto M. (2010). Staphylococcus colonization of the skin and antimicrobial peptides. Expert review of dermatology, 5(2), 183–195. https://doi.org/10.1586/edm.10.6
- Soumya, K. R., Philip, S., Sugathan, S., Mathew, J., & Radhakrishnan, E. K. (2017). Virulence factors associated with Coagulase Negative Staphylococci isolated from human infections. 3 Biotech, 7(2), 140. https://doi.org/10.1007/s13205-017-0753-2
- Giesbrecht, P., Kersten, T., Maidhof, H., & Wecke, J. (1998). Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiology and molecular biology reviews : MMBR, 62(4), 1371–1414.
- Mendoza-Olazarán, S., Morfín-Otero, R., Villarreal-Treviño, L., Rodríguez-Noriega, E., Llaca-Díaz, J., Camacho-Ortiz, A., González, G. M., Casillas-Vega, N., & Garza-González, E. (2015). Antibiotic Susceptibility of Biofilm Cells and Molecular Characterisation of Staphylococcus hominis Isolates from Blood. PloS one, 10(12), e0144684. https://doi.org/10.1371/journal.pone.0144684.
- Chaves, F., García-Alvarez, M., Sanz, F., Alba, C., & Otero, J. R. (2005). Nosocomial spread of a Staphylococcus hominis subsp. novobiosepticus strain causing sepsis in a neonatal intensive care unit. Journal of clinical microbiology, 43(9), 4877–4879. https://doi.org/10.1128/JCM.43.9.4877-4879.2005.
