Genes are basic units of inheritance that contain the genetic information necessary to determine the specific traits in an organism. Genes contain the instructions for producing proteins that perform various functions to keep the cell alive and functioning. Protein plays specific roles in the cell, contributing to the structure, function, and regulation of various biological processes. In order for a cell to function, it’s crucial that specific proteins are created at the right time.
This process of activation of genes to produce proteins is called gene expression. This activation leads to the creation of messenger RNA (mRNA), which acts as a template for building the proteins. Simply, gene expression is the flow of genetic information from gene to protein. Gene expression is a vital process in cells that ensures specific proteins are created at the right time and in the right quantities.
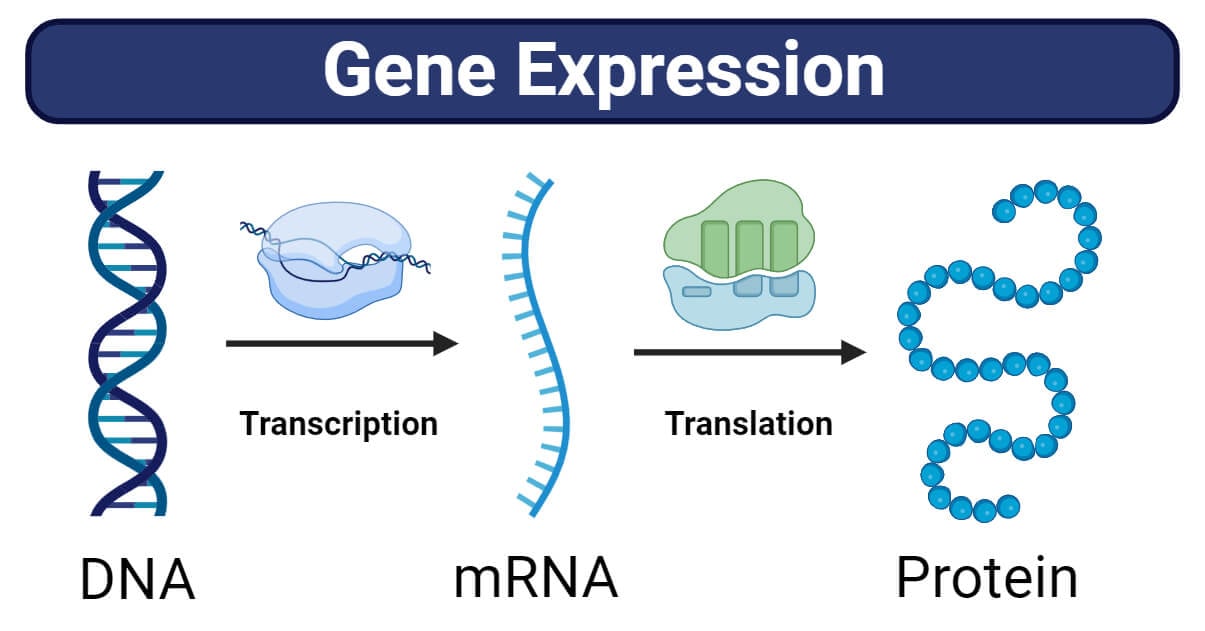
When a cell requires a specific protein, it activates the corresponding gene. This activation initiates a series of steps. First, an mRNA copy of the gene is produced through a process called transcription. The mRNA acts as a template for protein synthesis during translation. This process involves reading the mRNA sequence in triplets and assembling the corresponding amino acids to form a specific protein. The precise sequence of amino acids determines the structure and function of the protein.
Interesting Science Videos
Stages in Gene Expression
Gene expression is a biological process in which the genetic information contained within genes is translated into functional outcomes. This process includes two main stages: transcription and translation.
Transcription
Transcription is the first step in gene expression, where the genetic information encoded in DNA is transcribed into mRNA by an enzyme called RNA polymerase.
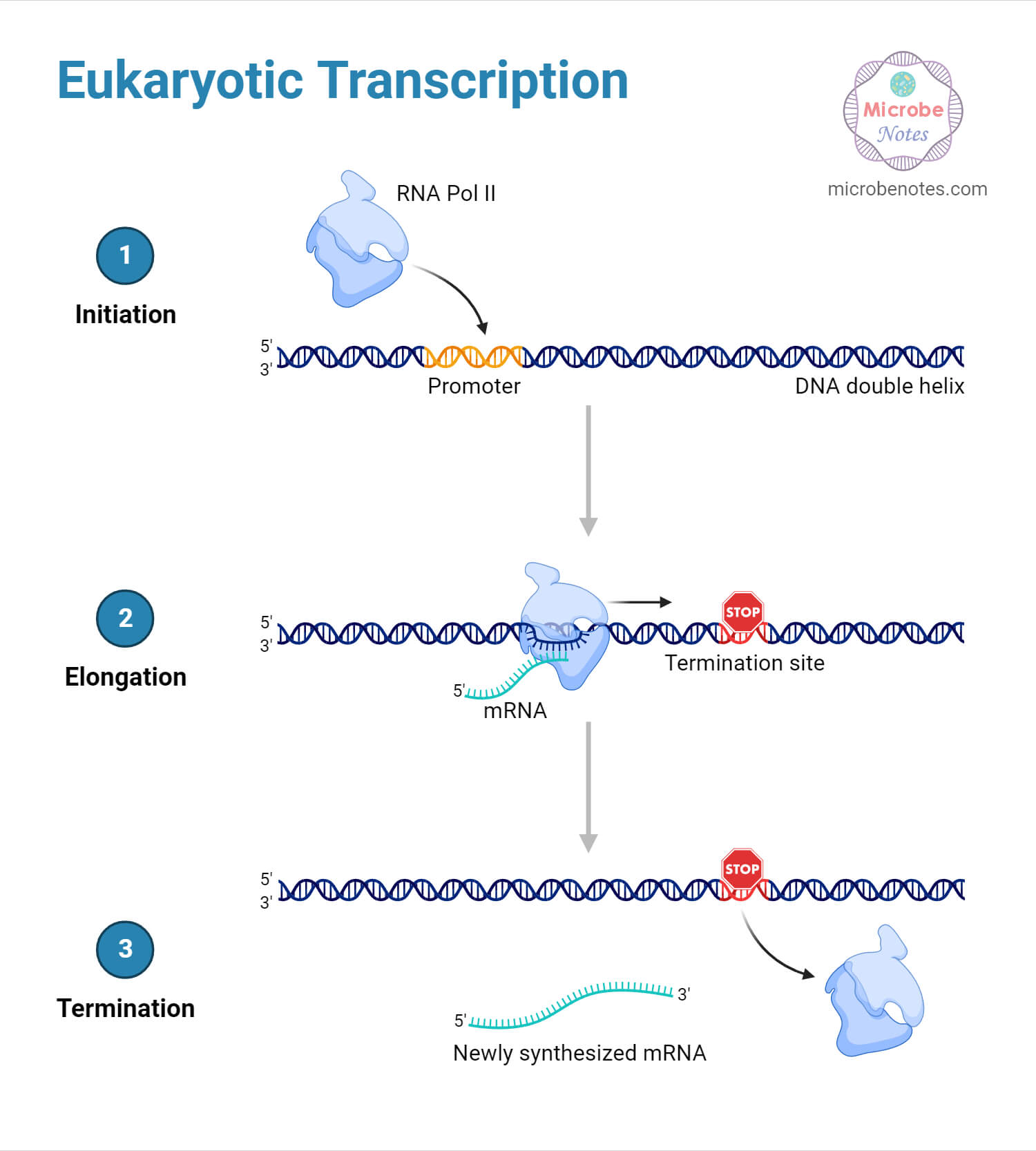
- The process of transcription starts with the binding of RNA polymerase to a specific region of the DNA molecule called the promoter. The promoter signals the starting point for transcription.
- RNA polymerase unwinds the DNA double helix near the promoter region. This allows one of the DNA strands to act as a template for the synthesis of mRNA.
- As RNA polymerase moves along the DNA template, it adds complementary RNA nucleotides to the growing mRNA chain. The RNA molecule is synthesized in the 5′ to 3′ direction, complementary to the DNA template strand.
- Transcription continues until a termination signal is reached in the DNA sequence. This signal causes RNA polymerase to separate from the DNA template and the mRNA molecule is released.
- In prokaryotes, transcription and translation occur in the same compartment.
- In eukaryotes, the initial RNA transcript (pre-mRNA) undergoes modifications before becoming mature mRNA. These modifications include capping, splicing, and adding a poly-A tail to the mRNA molecule.
- Once processed, mature mRNA molecules are transported from the cell nucleus into the cytoplasm, where they serve as templates for protein synthesis during translation.
Translation
Translation is the process that uses the genetic information encoded in mRNA to synthesize a specific sequence of amino acids, forming a functional protein.
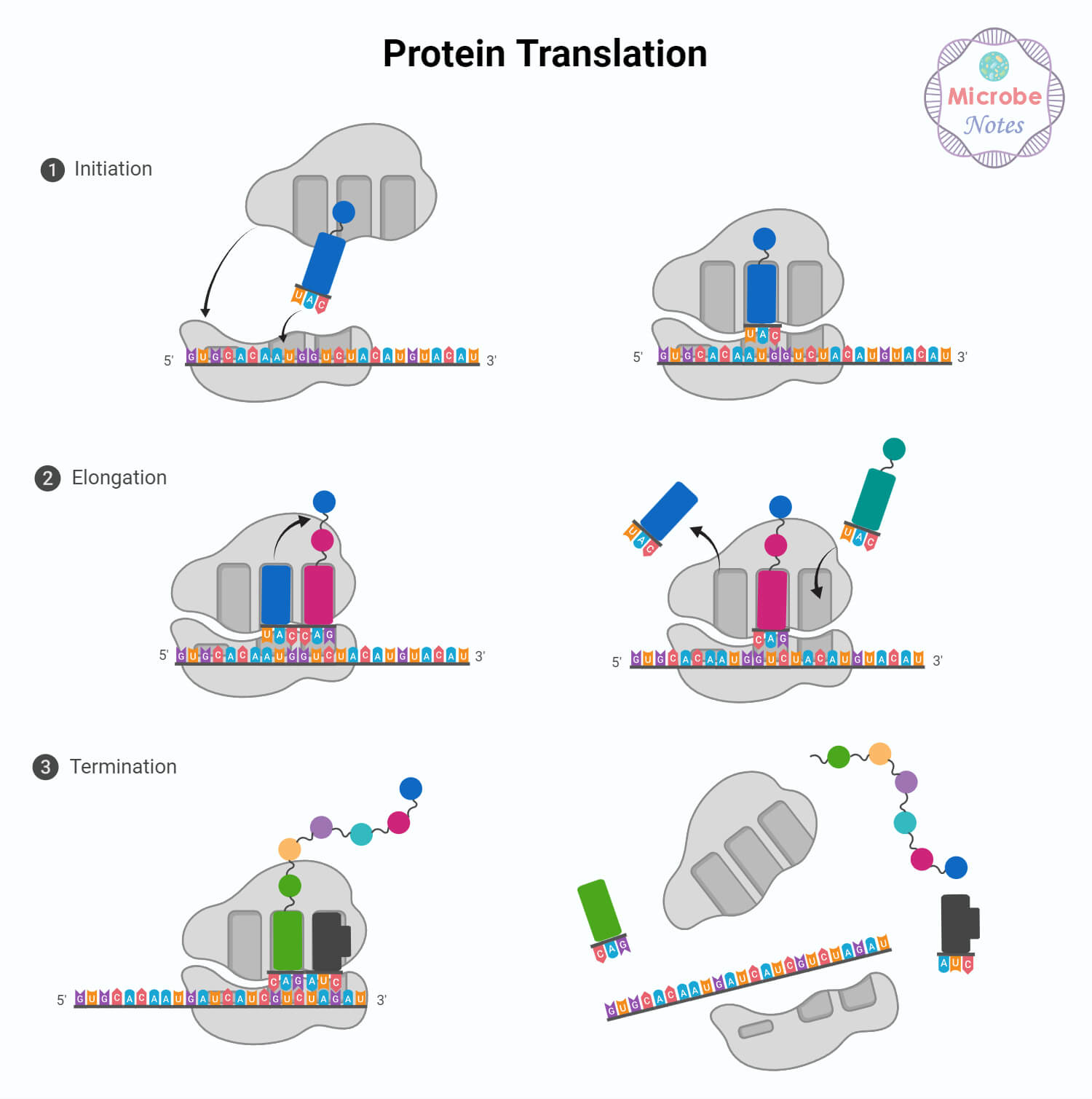
- Translation begins when the mRNA binds to the ribosome which recognizes the start codon (AUG) on the mRNA, marking the initiation point for translation.
- Transfer RNA (tRNA) molecules, each carrying a specific amino acid, match their anticodon sequences with the corresponding codons on the mRNA.
- During elongation, the ribosome moves along the mRNA, and each codon is matched with the appropriate tRNA anticodon. Amino acids carried by tRNAs are joined together in a specific sequence, forming a polypeptide chain.
- As the ribosome moves along the mRNA, this chain grows, creating the primary structure of the protein.
- Translation continues until a stop codon (UAA, UAG, or UGA) is encountered on the mRNA. Stop codons do not code for any amino acids; instead, they signal the end of the translation process.
- After translation, the newly synthesized protein may undergo various post-translational modifications. These modifications can include folding into a specific three-dimensional shape, addition of functional groups, or cleavage of specific segments to activate the protein.
Regulation of gene expression
Regulation of gene expression is a vital process that controls when and how much genes produce proteins. Gene expression is regulated to ensure that proteins are generated in specific cells at specific times.
Gene expression is regulated at multiple stages: transcription, RNA processing, translation, and post-translational modification.
1. Transcriptional regulation
Transcription regulation involves genetic interactions with specific DNA sites and epigenetic effects. Transcription factors interact with DNA, enhancing or inhibiting RNA polymerase binding. Epigenetic modifications, like DNA methylation and histone alterations, change gene accessibility without changing the DNA sequence.
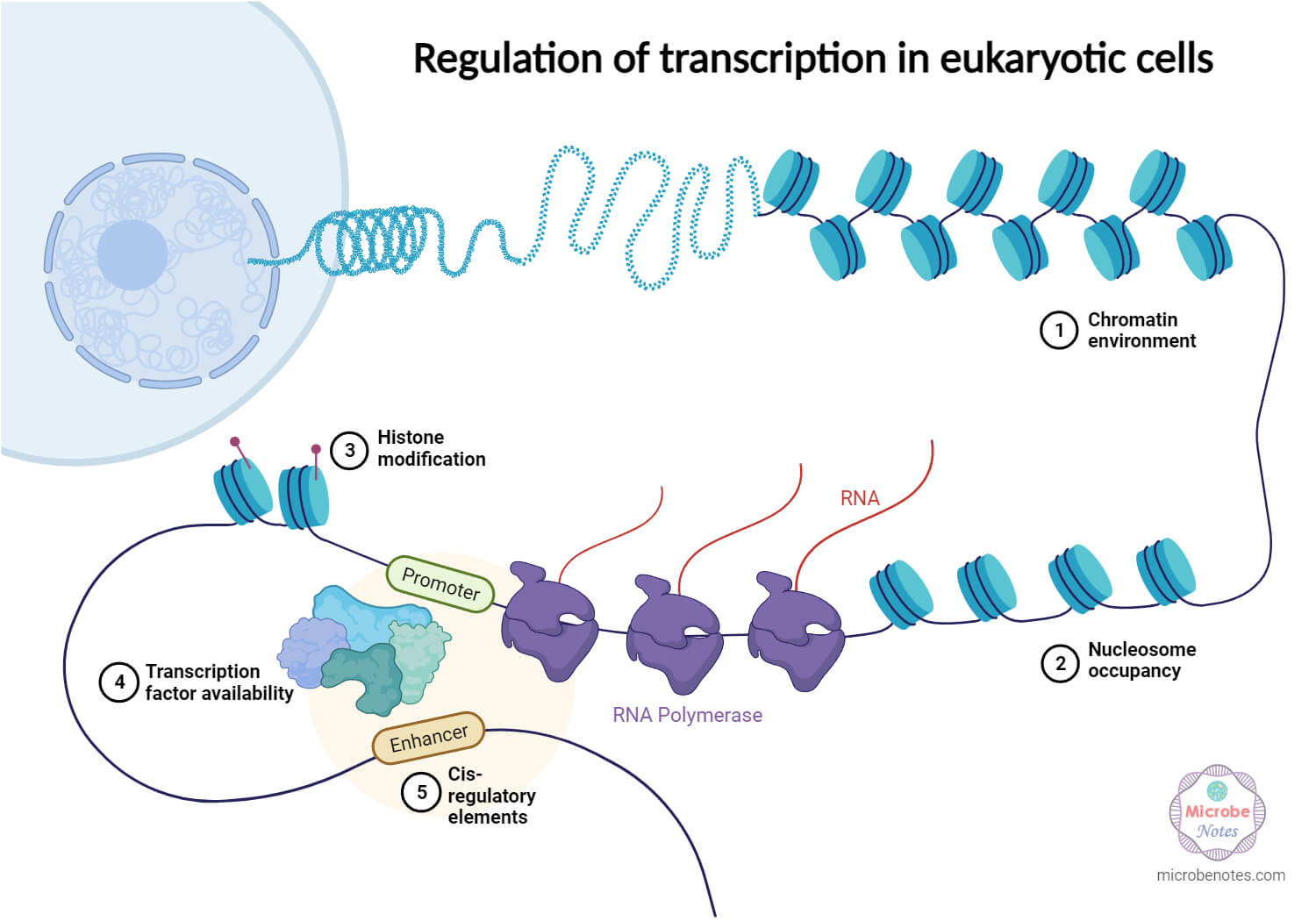
2. Post-transcriptional regulation
Gene expression can be regulated by post-transcriptional mRNA processing. In eukaryotes, transcribed mRNA undergoes splicing and other modifications that protect the ends of the RNA strand from degradation. Splicing removes non-coding segments called introns and joins the protein-coding regions called exons.
3. Translational regulation
Translation can be regulated by microRNAs that attach to a specific mRNA sequence, blocking translation initiation or causing mRNA degradation. Additionally, translational repressors can attach to RNA and interfere with translation initiation. Various other factors, including RNA-binding proteins and specific sequence elements in the mRNA, can also influence the process of translation.
4. Post-translational regulation
Translated polypeptides are processed to create functional proteins. Post-translational regulation alters the structure and function of proteins by adding or removing chemical groups like phosphorylation and acetylation, which affect protein activity and gene expression.
Methods for measuring gene expression
Measuring and quantifying gene expression is important for understanding biological processes. Some of the common methods and tools for measuring gene expression are:
- Northern Blotting is a hybridization-based method used to determine the size and quantity of specific RNA in a sample. It separates RNA by gel electrophoresis and transfers it to a nitrocellulose membrane, where it forms covalent bonds. Labeled probes complementary to the RNA transcript are used for hybridization. The method is simple and cost-effective, but it is time-consuming and has low throughput.
- Quantitative Polymerase Chain Reaction (qPCR) measures gene expression in real-time by converting mRNA to complementary DNA (cDNA) and amplifying it using PCR. Fluorescent labels are added, and the fluorescence is measured after each cycle, which provides quantitative data. This method is easy to use and has a relatively short assay time. However, it requires prior knowledge of the target sequence and has limited throughput.
- Microarray measures gene expression by hybridizing cDNA strands to quantify transcripts. Microarrays use oligonucleotides or cDNA attached to a chip surface. It allows high-throughput analysis. Despite its ability to measure a large number of transcripts, microarrays require separate preparation of control and test samples. Also, specialized equipment is necessary for data analysis.
- RNA-Seq directly sequences RNA and measures expression by counting sequences. RNA samples are processed into libraries, sequenced, and the data is analyzed to determine gene expression levels. It provides a higher range than microarrays and can detect various RNA types but it comes at a higher cost and requires more computational resources.
References
- Clancy, S. & Brown, W. (2008) Translation: DNA to mRNA to Protein. Nature Education 1(1):101. https://www.nature.com/scitable/topicpage/translation-dna-to-mrna-to-protein-393/
- Gene Expression (genome.gov)
- Gene Expression (nih.gov)
- Gene Expression | Learn Science at Scitable (nature.com)
- Gene Expression: Transcription, Splicing and Translation | Biology | JoVE
- Lewin B. (2000) Genes VII. Oxford University Press Inc., New York, USA.
- P, Surat. (2021, January 27). A Guide to Understanding Gene Expression. AZoLifeSciences. https://www.azolifesciences.com/article/A-Guide-to-Understanding-Gene-Expression.aspx.
- Parker, J. (2001). Gene Expression. Brenner’s Encyclopedia of Genetics, 187–189. doi:10.1016/b978-0-12-374984-0.00587-8
- Singh, K. P., Miaskowski, C., Dhruva, A. A., Flowers, E., & Kober, K. M. (2018). Mechanisms and Measurement of Changes in Gene Expression. Biological Research for Nursing, 20(4), 369-382. https://doi.org/10.1177/1099800418772161
- What is Gene Expression Analysis? | Bio-Rad

Simple and clear diagrammatic representation enabled easy understanding.
Thanks.
Very useful nots .thanks for nots