Translation involves translating the sequence of a messenger RNA (mRNA) molecule to a sequence of amino acids during protein synthesis.
It is the process in which ribosomes in the cytoplasm or ER synthesize proteins after the process of transcription of DNA to RNA.
Interesting Science Videos
The Ribosomes
- Ribosomes exist normally as separate subunits that are composed of protein and rRNA.
- The subunits come together to form a ribosome when they bind to an mRNA, near its 5’ end.
- On binding to an mRNA, the ribosome reads the nucleotide sequence from the 5’ to 3’ direction, synthesizing the corresponding protein from amino acids in an N-terminal (amino-terminal) to C-terminal (carboxyl terminal) direction.
- Ribosomes are located in the cytosol, either freely floating or associated with the endoplasmic reticulum.
- They serve to synthesize proteins.
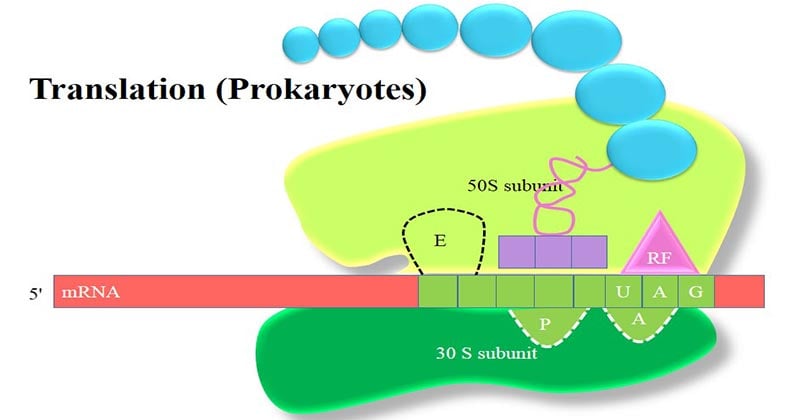
Ribosomal Sites for Protein Translation
Each prokaryotic ribosome, shown schematically, has three binding sites for tRNAs.
- The aminoacyl-tRNA binding site (or A site) is where, during elongation, the incoming aminoacyl-tRNA binds.
- The peptidyl-tRNA binding site (or P site) is where the tRNA linked to the growing polypeptide chain is bound.
- The exit site (or E site) is a binding site for tRNA following its role in translation and prior to its release from the ribosome.
All three sites (A, P and E) are formed by the rRNA molecules in the ribosome.
Translation Process
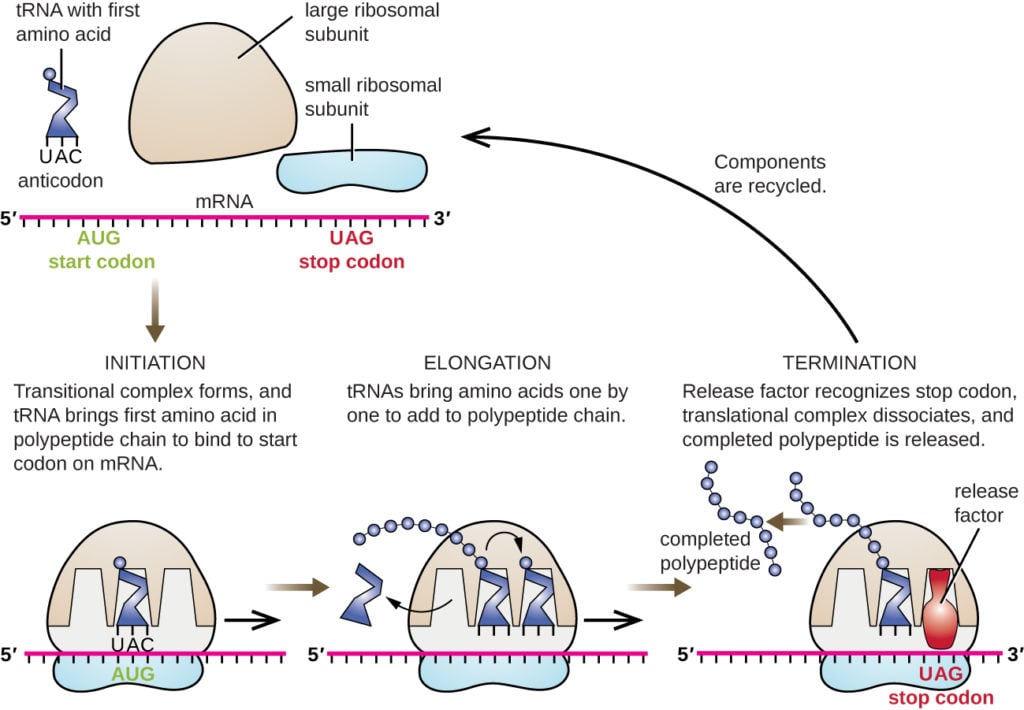
Protein synthesis (or translation) takes place in three stages:
- Initiation
- Elongation and
- Termination.
- During initiation, the mRNA–ribosome complex is formed and the first codon (always AUG) binds the first aminoacyltRNA (called initiator tRNA).
- During the elongation phase, the other codons are read sequentially and the polypeptide grows by addition of amino acids to its C-terminal end.
- This process continues until a termination codon (Stop codon), which does not have a corresponding aminoacyl-tRNA with which to base pair, is reached.
- At this point, protein synthesis ceases (termination phase) and the finished polypeptide is released from the ribosome.
Synthesis of aminoacyl-tRNA
- Synthesis of aminoacyl-tRNAs is crucially important for two reasons:
- Each amino acid must be covalently linked to a tRNA molecule in order to take part in protein synthesis, which depends upon the ‘adaptor’ function of tRNA to ensure that the correct amino acids are incorporated.
- The covalent bond that is formed between the amino acid and the tRNA is a high energy bond that enables the amino acid to react with the end of the growing polypeptide chain to form a new peptide bond.
For this reason, the synthesis of aminoacyl-tRNA is also referred to as amino acid activation.
- Each tRNA molecule has a cloverleaf secondary structure with the anticodon accessible at the end of the anticodon stem loop.
- During synthesis of the aminoacyl-tRNA, the amino acid is covalently bound to the A residue of the CCA sequence at the 3’ end.
- Each tRNA molecule carries only a single amino acid.
- The attachment of an amino acid to a tRNA is catalyzed by an enzyme called aminoacyl-tRNA synthetase.
- A separate aminoacyl-tRNA synthetase exists for every amino acid, making 20 synthetases in total.
The synthesis reaction occurs in two steps.
- The first step is the reaction of an amino acid and ATP to form an aminoacyl-adenylate (also known as aminoacyl-AMP).
- In the second step, without leaving the enzyme, the aminoacyl group of aminoacyl-AMP is transferred to the 3’ end of the tRNA molecule to form aminoacyl-tRNA
The overall reaction is:
Amino acid + ATP + tRNA → aminoacyl-tRNA + AMP + PPi
Initiation of Protein Synthesis
- The first codon translated in all mRNAs is the start codon or initiation codon, AUG which codes for methionine.
- Two different tRNAs are used for the two types of AUG codon; tRNAfMet is used for the initiation codon and is called the initiator tRNA whereas tRNAm Met is used for internal AUG codons.
- In prokaryotes the first amino acid of a new protein is N-formylmethionine (abbreviated fMet). Hence the aminoacyl-tRNA used in initiation is fMet-tRNAfMet.
- A short sequence rich in purines (5’-AGGAGGU-3’), called the Shine–Dalgarno sequence, lies 5’ to the AUG initiation codon and is complementary to part of the 16S rRNA in the small ribosomal subunit.
- Therefore this is the binding site for the 30S ribosomal subunit which then migrates in a 3’ direction along the mRNA until it encounters the AUG initiation codon.
- Initiation of protein synthesis requires proteins called initiation factors (IFs).
- In prokaryotes, three initiation factors (IF-1, IF-2 and IF-3) are essential.
- Because of the complexity of the process, the exact order of binding of IF-1, IF-2, IF-3, fMet-tRNAf is controversial.
Steps Involved
- Initiation begins with the binding of IF-1 and IF-3 to the small (30S) ribosomal subunit.
- Their role is to stop the 30S subunit binding to the 50S subunit in the absence of mRNA and fMet-tRNAf Met which would result in a nonfunctional ribosome.
- The small subunit then binds to the mRNA via the Shine–Dalgarno sequence and moves 3’ along the mRNA until it locates the AUG initiation codon.
- The initiator tRNA charged with N-formylmethionine and in a complex with IF-2 and GTP (fMet-tRNAfMet/IF-2/GTP) now binds.
- IF-3 is released.
- The complex of mRNA, fMet-tRNAf Met, IF-1, IF-2 and the 30S ribosomal subunit is called the 30S initiation complex.
- The large (50S) ribosomal subunit now binds, with the release of IF-1 and IF-2 and hydrolysis of GTP, to form a 70S initiation complex.
Elongation of Protein Synthesis
- At the start of the first round of elongation, the initiation codon (AUG) is positioned in the P site with fMet-tRNAfMet bound to it via codon–anticodon base pairing.
- The next codon in the mRNA is positioned in the A site.
- Elongation of the polypeptide chain occurs in three steps called the elongation cycle, namely aminoacyl-tRNA binding, peptide bond formation and translocation:
Aminoacyl-tRNA binding
- The corresponding aminoacyl-tRNA for the second codon binds to the A site via codon–anticodon interaction.
- Binding of the aminoacyl-tRNA requires elongation factor EF-Tu and GTP which bind as an aminoacyl-tRNA/EF-Tu/GTP complex.
- Following binding, the GTP is hydrolyzed and the EF-Tu is released, now bound to GDP.
- Before the EF-Tu molecule can catalyze the binding of another charged tRNA to the ribosome, it must be regenerated by a process involving another elongation factor, EF-Ts.
This regeneration is called the EF-Tu–EF-Ts exchange cycle.
- First, EF-Ts binds to EF-Tu and displaces the GDP. Then GTP binds to the EF-Tu and displaces EF-Ts. The EF-Tu-GTP is now ready to take part in another round of elongation.
Peptide bond formation
- The second step, peptide bond formation, is catalyzed by peptidyl transferase.
- In this reaction the carboxyl end of the amino acid bound to the tRNA in the P site is uncoupled from the tRNA and becomes joined by a peptide bond to the amino group of the amino acid linked to the tRNA in the A site.
Translocation
- In the third step, a complex of elongation factor EF-G (also called translocase) and GTP (i.e. EF-G/GTP) binds to the ribosome.
- Three concerted movements now occur, collectively called translocation:
- the deacylated tRNA moves from the P site to the E site
- the dipeptidyl-tRNA in the A site moves to the P site, and
- the ribosome moves along the mRNA (5’ to 3’) by three nucleotides to place the next codon in the A site.
- During the translocation events, GTP is hydrolyzed to GDP and inorganic phosphate, and EF-G is released ready to bind more GTP for another round of elongation.
- After translocation, the A site is empty and ready to receive the next aminoacyltRNA.
- The A site and the E site cannot be occupied simultaneously. Thus the deacylated tRNA is released from the E site before the next aminoacyl-tRNA binds to the A site to start a new round of elongation.
- Elongation continues, adding one amino acid to the C-terminal end of the growing polypeptide for each codon that is read, with the peptidyl-tRNA moving back and forth from the P site to the A site as it grows.
Termination of Protein Synthesis
- Eventually, one of three termination codons (also called Stop codons) becomes positioned in the A site. These are UAG, UAA and UGA.
- Unlike other codons, prokaryotic cells do not contain aminoacyl-tRNAs complementary to
- Stop codons. Instead, one of two release factors (RF-1 and RF-2) binds instead.
- RF-1 recognizes UAA and UAG whereas RF-2 recognizes UAA and UGA. A third release factor, RF-3, is also needed to assist RF-1 or RF-2 interaction with the ribosome. Thus either RF-1 + RF-3 or RF-2 + RF-3 bind depending on the exact termination codon in the A site.
- RF-1 (or RF-2) binds at or near the A site whereas RF-3/GTP binds elsewhere on the ribosome.
- The release factors cause the peptidyl transferase activity to transfer the polypeptide to a water molecule instead of to aminoacyl-tRNA, effectively cleaving the bond between the polypeptide and tRNA in the P site.
The free polypeptide now leaves the ribosome, followed by the mRNA and free tRNA, and the ribosome dissociates into 30S and 50S subunits ready to start translation again.
References
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
- Madigan, M. T., Martinko, J. M., Bender, K. S., Buckley, D. H., & Stahl, D. A. (2015). Brock biology of microorganisms (Fourteenth edition.). Boston: Pearson.

Thank you so much for your well explainatory notes.. It really helped me a lot
nice explaination
Tq Sir it’s very useful nots
The notes were really helping
Great article, you SAVED me after looking at so so so much information on my native language that was convoluted and mixed with shady websites. Finally I found the perfect page to look at prokaryotic translation. God bless
Sir/madam can you send me the all notes/notes link in the whatsApp?