Northern blot is a technique based on the principle of blotting for the analysis of specific RNA in a complex mixture.
- The technique is a modified version of the Southern Blotting, which was discovered for the analysis of DNA sequences.
- The detection of certain sequences of nucleic acids extracted from different types of biological samples is essential in molecular biology, which makes blotting techniques imperative in the field.
- The principle is identical to southern blotting except for the probes used for the detection as northern blotting detects RNA sequences.
- This technique provides information about the length of the RNA sequences and the presence of variations in the sequence.
- Even though the technique is primarily focused on the identification of RNA sequences, it has also been used for the quantification of RNA sequences.
- Since the discovery of the technique, several modifications have been made in the technique for the analysis of mRNAs, pre-mRNAs, and short RNAs.
- Northern blotting was employed as the primary technique for the analysis of RNA fragments for a long time; however, new, more convenient, and cost-effective techniques like RT-PCR have slowly replaced the technique.
Interesting Science Videos
Principle of Northern Blot
- The principle of the northern blot is the same as all other blotting technique that is based on the transfer of biomolecules from one membrane to another.
- The RNA samples are separated on gels according to their size by gel electrophoresis. Since RNAs are single-stranded, these can form secondary structures by intermolecular base pairing. The electrophoretic separation of the RNA segments is thus performed under denaturing conditions.
- The separated RNA fragments are then transferred to a nylon membrane. Nitrocellulose membrane is not used as RNA doesn’t bind effectively to the membrane.
- The transferred segments are immobilized onto the membrane by fixing agents. The RNA fragments on the membrane are detected by the addition of a labeled probe complementary to the RNA sequences present on the membrane.
- The hybridization forms the basis of the detection of RNA as the specificity of hybridization between the probe, and the RNA allows the accurate identification of the segments.
- Northern blot utilizes size-dependent separation of RNA segments and thus can be used to determine the sizes of the transcripts.
Requirements
Equipment
- Agarose Gel cast
- Power Supply
- Microwave
- Centrifuge
- Heating block
- UV crosslinker
- Hybridization oven
- Hybridization vessels
- Vials
- Forceps
- Pipettes
- Glass tubes
Materials
- Agarose gel
- Sodium citrate
- Ethylenediaminetetraacetic acid disodium salt dehydrate
- NaOH
- HCl
- Formaldehyde
- Glycerol
- Ethidium bromide
- Bromophenol Blue
- RNA ladder
- MgCl2
- NaCl
- Polyvinylpyrrolidone
- Bovine Serum Albumin
- SDS
- NaH2PO4
- Tris-HCl
- Triton-X
- DTT
- Taq buffer
- Taq polymerase
Procedure/Steps of Northern Blot
a. Separation of RNA on a denaturing gel
- The RNA gel solution is prepared by adding formaldehyde to the agarose solution.
- The cast is assembled, and the prepared denaturing gel is poured into the cast. As the gel begins to set, a comb with appropriate teeth is added to form wells.
- Once the gel is set, the comb is removed, and the gel is equilibrated with a running buffer for 30 minutes before running.
- 15 µg RNA sample is mixed with an equal volume of RNA loading buffer. Three µg of RNA markers are added in the same volume of RNA loading buffer.
- The samples are incubated at 65°C on a heating block for about 12-15 minutes.
- The samples are loaded to the equilibrated gel, and the first row of wells is filled with RNA markers.
- The gel is then run at 125V for about 3 hours.
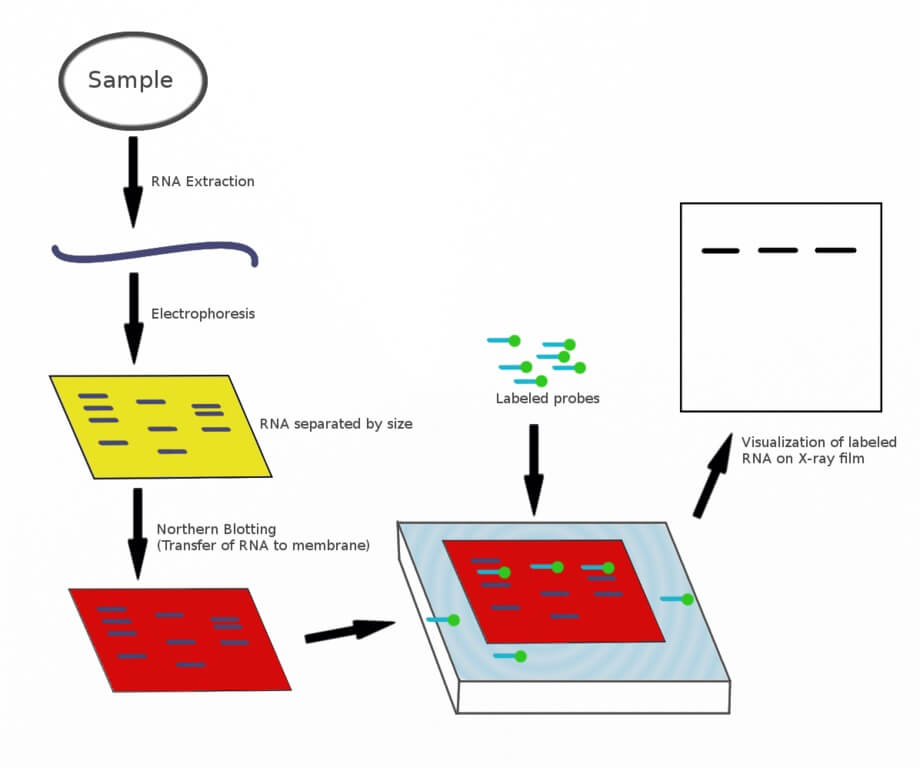
Image Source: Ilewieszoośmiornicach (Wikimedia).
b. Transfer of RNA from gel to the nylon membrane
- A nylon membrane is cut that is larger than the size of the denaturing gel, and a filter paper with the same size as the nylon membrane is also prepared.
- Once the electrophoresis process is complete, the RNA gel is removed from the tank and rinsed with water.
- An oblong sponge that is slightly larger than the gel is placed on a glass dish, and the dish is filled with SSC to a point so as to leave the soaked sponge about half-submerged in the buffer.
- A few pieces of Whatman 3mm papers are placed on top of the sponge and are wetted with SSC buffer.
- The gel is then placed on top of the filter paper and squeezed out to remove air bubbles by rolling a glass pipette over the surface.
- The nylon membrane prepared is wetted with distilled water on an RNase-free dish for about 5 minutes.
- The wetted membrane is placed on the surface of the gel while avoiding any air bubbles formation.
- The surface is further flooded with SSC, and a few more filter papers are placed on top of the membrane.
- A glass plate is placed on top of the structure in order to hold everything in place. The structure is left overnight to obtain an effective transfer.
c. Immobilization
- Once the transfer is complete, the gel is removed and rinsed with SSC, and allowed to dry.
- The membrane is placed between two pieces of filter paper and baked in a vacuum oven at 80°C for 2 hours.
- In some cases, the membrane can be wrapped in a UV transparent plastic wrap and irradiates for an appropriate time on a UV transilluminator.
d. Hybridization
- The DNA or RNA probes to be used are to be labeled to a specific activity of >108 dpm/µg, and unincorporated nucleotides are to be removed.
- The membrane carrying the immobilized RNA is wetted with SSC.
- The membrane is placed in a hybridization tube with the RNA-side-up, and 1 ml of formaldehyde solution is added.
- The tube is placed in the hybridization oven and incubated at 42°C for 3 hours.
- If the probe used is double-stranded, it is denatured by heating in a water bath or incubator for 10 minutes at 100°C.
- The desired volume of the probe is pipette into the hybridization tube and further incubated at 42°C.
- The solution is poured off, and the membrane is washed with a wash solution. The membrane is then observed under autoradiography.
Result Interpretation of Northern Blot
The RNA bands are observed under radiography in the form of bands. The distance of the bands from the markers can be used to determine the length and semi quantification of the RNA fragments.
Applications of Northern Blot
- The technique can be used for the identification and separation of RNA fragments collected from different biological sources.
- Northern blotting is used as a sensitive test for the detection of transcription of DNA fragments that are to be used as a probe in Southern Blotting.
- It also allows the detection and quantification of specific mRNAs from different tissues and different living organisms.
- Northern blotting is used as a tool for gene expression studies related to overexpression of cancer-causing genes, and gene expression during transplant rejects.
- Northern blotting has been used as a molecular tool for the diagnosis of diseases like Crohn’s disease.
- The process is used as a method for the detection of viral microRNAs that play important roles in viral infection.
Limitations of Northern Blot
- Northern blotting has a lower sensitivity as compared to other modern techniques like RT-PCR and nuclease protection assays.
- The method requires a large amount of sample RNA, and these should be of high quality.
- The technique is time-consuming and complex, especially in cases where multiple probes are to be added.
References
- He, Shan L, and Rachel Green. “Northern blotting.” Methods in enzymology vol. 530 (2013): 75-87. doi:10.1016/B978-0-12-420037-1.00003-8
- Josefsen K, Nielsen H. Northern blotting analysis. Methods Mol Biol. 2011;703:87-105. doi: 10.1007/978-1-59745-248-9_7. PMID: 21125485.
- Blevins T. Northern blotting techniques for small RNAs. Methods Mol Biol. 2010;631:87-107. doi: 10.1007/978-1-60761-646-7_9. PMID: 20204871.
- Mishima, Eikan et al. “Immuno-Northern Blotting: Detection of RNA Modifications by Using Antibodies against Modified Nucleosides.” PloS one vol. 10,11 e0143756. 25 Nov. 2015, doi:10.1371/journal.pone.0143756
- Koscianska, Edyta et al. “Northern blotting analysis of microRNAs, their precursors and RNA interference triggers.” BMC molecular biology vol. 12 14. 11 Apr. 2011, doi:10.1186/1471-2199-12-14
- Brown T, Mackey K. Analysis of RNA by northern blot hybridization. Curr Protoc Hum Genet. 2001 Nov;Appendix 3:Appendix 3K. doi: 10.1002/0471142905.hga03ks30. PMID: 18428227.
Sources
- https://www.routledgehandbooks.com/doi/10.1201/9780203507438.ch3 – 11%
- https://en.wikipedia.org/wiki/Northern_blot – 5%
- https://www.sciencedirect.com/topics/medicine-and-dentistry/northern-blotting – 4%
- https://microbenotes.com/southern-blot/ – 3%
- https://findanyanswer.com/what-is-southern-blotting-technique – 2%
- https://www.sciencedirect.com/science/article/pii/B9780124200371000038 – 1%
- https://www.thermofisher.com/us/en/home/references/ambion-tech-support/northern-analysis/tech-notes/strategies-for-detecting-mrna.html – 1%
- https://nanocellect.com/blog/rna-sequencing-analysis-everything-you-need-to-know/ – 1%
- https://pubmed.ncbi.nlm.nih.gov/18428227/ – 1%
- https://www.biotechniques.com/biochemistry/south-north-east-and-west-ern-the-story-of-how-the-western-blot-came-into-being/ – 1%
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7515567/ – <1%

Seria legal se estivesse um video animado da tecnica so para que o entendimento seje mais especificos aos leigos no assunto.
👍👍