Protein Synthesis involves translating the sequence of a messenger RNA (mRNA) molecule to a sequence of amino acids during protein synthesis. It is the process in which ribosomes in the cytoplasm or ER synthesize proteins after the process of transcription of DNA to RNA.
Interesting Science Videos
The Ribosomes
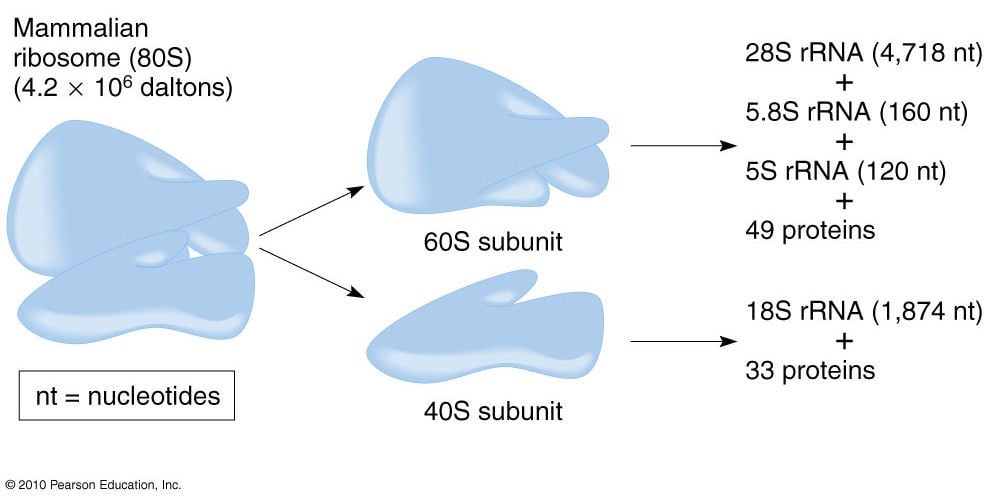
- Ribosomes exist normally as separate subunits that are composed of protein and rRNA.
- Eukaryotic ribosomes are larger (the 80S) and more complex than prokaryotic ribosomes (70S).
- The subunits come together to form a ribosome when they bind to an mRNA, near its 5’ end.
- On binding to an mRNA, the ribosome reads the nucleotide sequence from the 5’ to 3’ direction, synthesizing the corresponding protein from amino acids in an N-terminal (amino-terminal) to C-terminal (carboxyl-terminal) direction.
- Ribosomes are located in the cytosol, either freely floating or associated with the endoplasmic reticulum.
- They serve to synthesize proteins.
Ribosomal Sites for Protein Synthesis
Each prokaryotic ribosome, shown schematically, has three binding sites for tRNAs.
- The aminoacyl-tRNA binding site (or A site) is where, during elongation, the incoming aminoacyl-tRNA binds.
- The peptidyl-tRNA binding site (or P site) is where the tRNA linked to the growing polypeptide chain is bound.
- The exit site (or E site) is a binding site for tRNA following its role in translation and prior to its release from the ribosome.
All three sites (A, P, and E) are formed by the rRNA molecules in the ribosome.
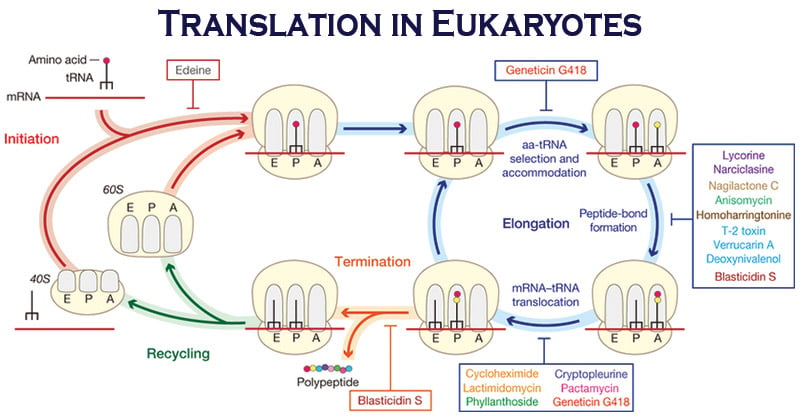
Protein Synthesis Process
The overall mechanism of protein synthesis in eukaryotes is basically the same as in prokaryotes.
However, there are some significant differences:
- Whereas a prokaryotic ribosome has a sedimentation coefficient of the 70S and subunits of 30S and 50S, a eukaryotic ribosome has a sedimentation coefficient of 80S with subunits of 40S and 60S.
- The composition of eukaryotic ribosomal subunits is also more complex than prokaryotic subunits but the function of each subunit is essentially the same as in prokaryotes.
- In eukaryotes, each mRNA is monocistronic that is, discounting any subsequent post-translational cleavage reactions that may occur; the mRNA encodes a single protein. In prokaryotes, many mRNAs are polycistronic that is they encode several proteins. Each coding sequence in a prokaryotic mRNA has its own initiation and termination codons.
- Initiation of protein synthesis in eukaryotes requires at least nine distinct eukaryotic initiation factors (eIFs) compared with the three initiation factors (IFs) in prokaryotes.
- In eukaryotes, the initiating amino acid is methionine, not N-formylmethionine as in prokaryotes.
- As in prokaryotes, a special initiator tRNA is required for initiation and is distinct from the tRNA that recognizes and binds to codons for methionine at internal positions in the mRNA. When charged with methionine ready to begin initiation, this is known as Met-tRNAimet
- The main difference between initiation of translation in prokaryotes and eukaryotes is that in bacteria, a Shine–Dalgarno sequence lies 5’ to the AUG initiation codon and is the binding site for the 30S ribosomal subunit.
- In contrast, most eukaryotic mRNAs do not contain Shine–Dalgarno sequences. Instead, a 40S ribosomal subunit attaches at the 5’ end of the mRNA and moves downstream (i.e. in a 5’ to 3’ direction) until it finds the AUG initiation codon. This process is called scanning.
- Prokaryotic translation requires no helicase, presumably because protein synthesis in bacteria can start even as the mRNA is still being synthesized whereas, in eukaryotes, transcription in the nucleus and translation in the cytoplasm are separate events that allow time for mRNA secondary structure to form.
Protein synthesis (or translation) takes place in three stages:
- Initiation
- Elongation and
- Termination
Initiation of Protein Synthesis
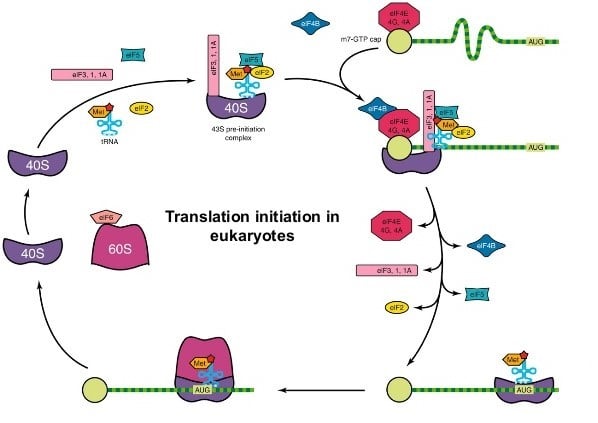
- The first step is the formation of a pre-initiation complex consisting of the 40S small ribosomal subunit, Met-tRNAimet, eIF-2, and GTP.
- The pre-initiation complex binds to the 5’ end of the eukaryotic mRNA, a step that requires eIF-4F (also called cap-binding complex) and eIF-3.
- The eIF-4F complex consists of eIF-4A, eIF-4E, and eIF-4G; eIF-4E binds to the 5’ cap on the mRNA whilst eIF-4G interacts with the poly (A) binding protein on the poly (A) tail.
- The eIF-4A is an ATP-dependent RNA helicase that unwinds any secondary structures in the mRNA, preparing it for translation.
- The complex then moves along the mRNA in a 5’ to 3’ direction until it locates the AUG initiation codon (i.e. scanning).
- The 5’ untranslated regions of eukaryotic mRNAs vary in length but can be several hundred nucleotides long and may contain secondary structures such as hairpin loops. These secondary structures are probably removed by initiation factors of the scanning complex.
- The initiation codon is usually recognizable because it is often (but not always) contained in a short sequence called the Kozak consensus (5’-ACCAUGG-3’).
- Once the complex is positioned over the initiation codon, the 60S large ribosomal subunit binds to form an 80S initiation complex, a step that requires the hydrolysis of GTP and leads to the release of several initiation factors.
Elongation of Protein Synthesis
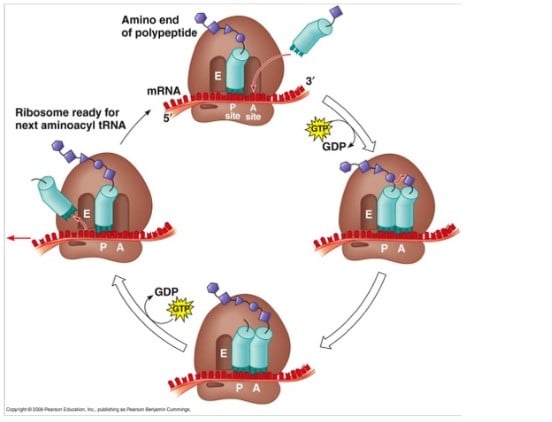
- Elongation depends on eukaryotic elongation factors.
- Three elongation factors, eEF-1A, eEF-IB, and eEF-2, are involved which have similar functions to their prokaryotic counterparts EF-Tu, EF-Ts and EF-G.
- At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis.
- The initiator tRNA occupies the P site in the ribosome, and the A site is ready to receive an aminoacyl-tRNA.
- During chain elongation, each additional amino acid is added to the nascent polypeptide chain in a three-step microcycle.
- The steps in this microcycle are:
- Positioning the correct aminoacyl-tRNA in the A site of the ribosome,
- Forming the peptide bond and
- Shifting the mRNA by one codon relative to the ribosome.
- Although most codons encode the same amino acids in both prokaryotes and eukaryotes, the mRNAs synthesized within the organelles of some eukaryotes use a variant of the genetic code.
- During elongation in bacteria, the deacylated tRNA in the P site moves to the E site prior to leaving the ribosome. In contrast, although the situation is still not completely clear, in eukaryotes the deacylated tRNA appears to be ejected directly from the ribosome.
Termination of Protein Synthesis
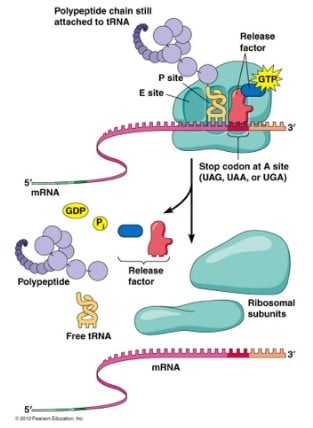
- Termination of elongation depends on eukaryotic release factors.
- In eukaryotes, eukaryotic release factor eRF-1 recognizes all three termination codons (UAA, UAG, and UGA) and, with the help of protein eRF-3, terminates translation.
- Upon termination, the ribosome is disassembled and the completed polypeptide is released.
References
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
- Madigan, M. T., Martinko, J. M., Bender, K. S., Buckley, D. H., & Stahl, D. A. (2015). Brock biology of microorganisms (Fourteenth edition.). Boston: Pearson.
- https://en.wikipedia.org/wiki/Eukaryotic_translation

Would you give me a summary of translation in eukaryotes
The information got is clear and broard although the diagrams are few compared to how needful they are in this process