Protein Synthesis is a process of synthesizing proteins in a chain of amino acids known as polypeptides. It is the second part of the central dogma in genetics.
- It takes place in the ribosomes found in the cytosol or those attached to the rough endoplasmic reticulum.
- The functions of the ribosome are to read the sequence of the codons in mRNA and the tRNA molecules that transfer or transport or bring the amino acids to the ribosomes in the correct sequence. However, other molecules are also involved in the process of translation such as various enzymatic factors.
- The translation process involves reading the genetic code in mRNA to make proteins.
- The entire translation process can be summarized into three phases: Initiation, elongation, and termination.
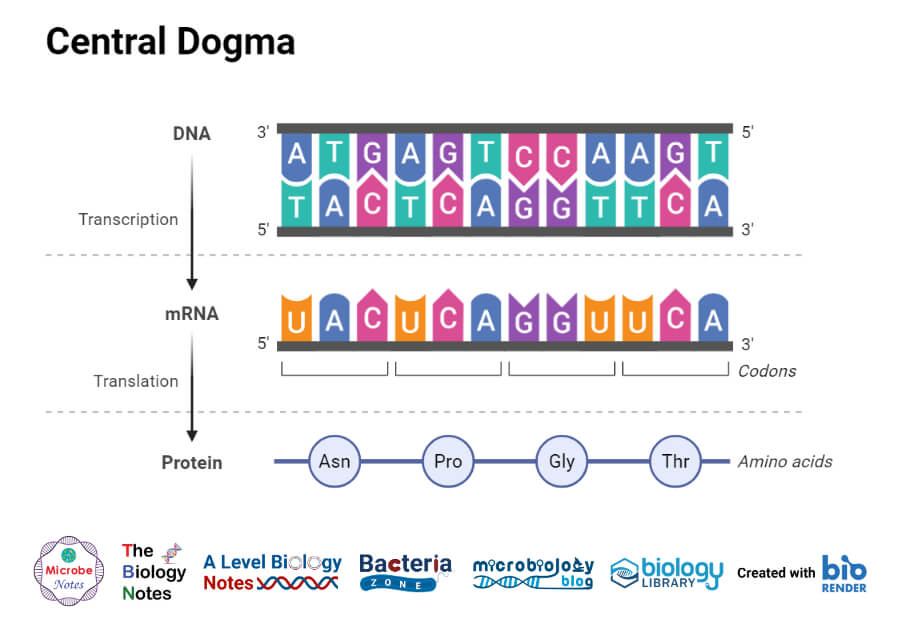
Figure: Central Dogma.
Interesting Science Videos
Protein Synthesis Machinery
The translation process is aided by two major factors: A translator – this is the molecule that conducts the translation; substrate – this is where the mRNA is translated into a new protein (translator desk). The translation process is guided by machinery composed of:
Ribosomes
- Ribosomes are made of ribosomal RNA (rRNA) and proteins, and therefore they are also named ribozymes because the rRNA has enzymatic activity. the rRNA has the peptidyl transferase activity that bonds the amino acids.
- The ribosomes have two subunits of rRNA and proteins, a large subunit with three active sites (E, P, A) which are critical for the catalytic activity of ribosomes.
Transfer RNA (tRNA)
- Each tRNA has an anticodon for the amino acid codon it carries which are complementary to each other. For example; Lysine is coded by AAG, and therefore the anticodon that will be carried by tRNA will be UUC, therefore when the codon AAG appears, an anticodon UUC of tRNA will bind to it temporarily.
- When tRNA is bound to mRNA, the tRNA then releases its amino acid. rRNA then helps to form bonds between the amino acids as they are transported to the ribosomes one by one, thus creating a polypeptide chain. The polypeptide chain keeps growing until it reaches a stop codon.
Protein Synthesis enzymes and functions
- Peptidyl transferase is the main enzyme used in Translation. It is found in the ribosomes with an enzymatic activity that catalyzes the formation of a covalent peptide bond between the adjacent amino acids.
- The enzyme’s activity is to form peptide bonds between adjacent amino acids using tRNAs during translation.
- The enzyme’s activity uses two substrates of which one has the growing peptide chain and the other bears the amino acid that is added to the chain.
- It is located in the large subunit of the ribosomes and therefore, the primary function of peptidyl transferase is to catalyze the addition of amino acid residues allowing the polypeptide chain to grow.
- The peptidyl transferase enzyme is entirely made up of RNA and its mechanism is mediated by ribosomal RNA (rRNA), which is a ribozyme, made up of ribonucleotides.
- In prokaryotes, the 23S subunit contains the peptidyl transferase between the A-site and the O-site of tRNA while in eukaryotes, it is found in the 28S subunit.
Overview of the Protein Synthesis
- The ribosomal translation is initiated when the ribosomes recognize the starting point of mRNA, where it binds a molecule of tRNA that bears a single amino acid.
- In prokaryotes, the initial amino acid in N-formylmethionine. during elongation, the second amino acid is linked to the first one.
- The ribosome then shifts its position on the mRNA and repeats the elongation cycle.
- When the elongation process reaches the stop codon, the amino acid chain folds spontaneously to form a protein.
- The ribosomes then split into two subunits, but later rejoin before another mRNA is translated.
- Protein synthesis is facilitated by several catalytic proteins which include initiation, elongation, termination factors, and guanosine triphosphates (GTP).
- GTP is a molecule that releases energy when converted into guanosine diphosphate (GDP).
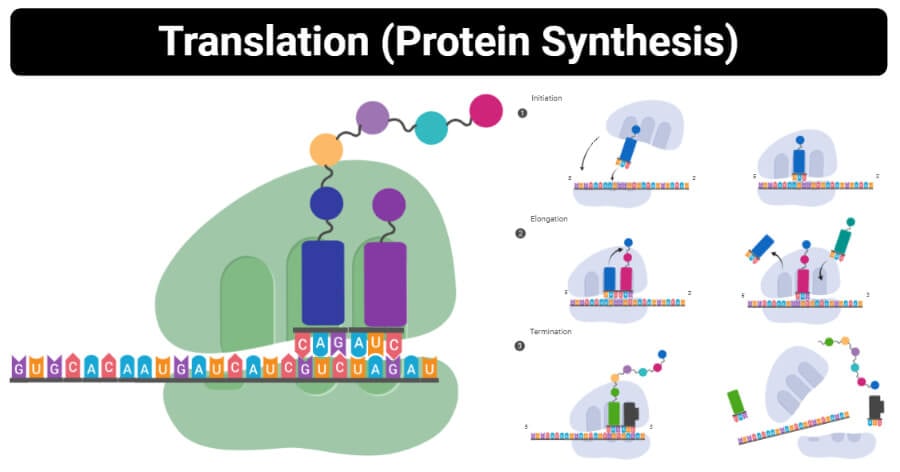
Protein Synthesis Steps /Process in Details
Translation Initiation
- Protein synthesis initiation is triggered by the presence of several initiation factors IF1, IF2, and IF3, including mRNA, ribosomes, tRNA.
- The small subunit binds to the upstream on the 5′ end at the start of mRNA. The ribosome scans the mRNA in the 5′ to 3′ direction until it encounters the start codon (AUG or GUG or UUG). When either of these start codons is present, it is recognized by the initiator fMet-tRNA (N-formylMet-tRNA). This initiator factor carries the methionine (Met) which binds to the P site on the ribosome.
- This synthesizes the first amino acid polypeptide known as N-formylmethionine. The initiator fMet-tRNA has a normal methionine anticodon therefore it inserts the N-formylmethionine. This means that methionine is the first amino acid that is added and appears in the chain.
- Generally, there are three steps in the initiation process of translation;
- Initiation of the binding of mRNA to the small ribosome subunit (the 30S), stimulating the initiator factor IF3. this dissociates the ribosomal subunits into two.
- The initiator factor IF2 then binds to the Guanine-triphosphate (GTP) and to the initiator fMet-tRNA to the P-site of the ribosomes.
- A ribosomal protein splits the GTP that is bound to IF2 thus helping in driving the assembly of the two ribosomal subunits. The IF3 and IF2 are released.
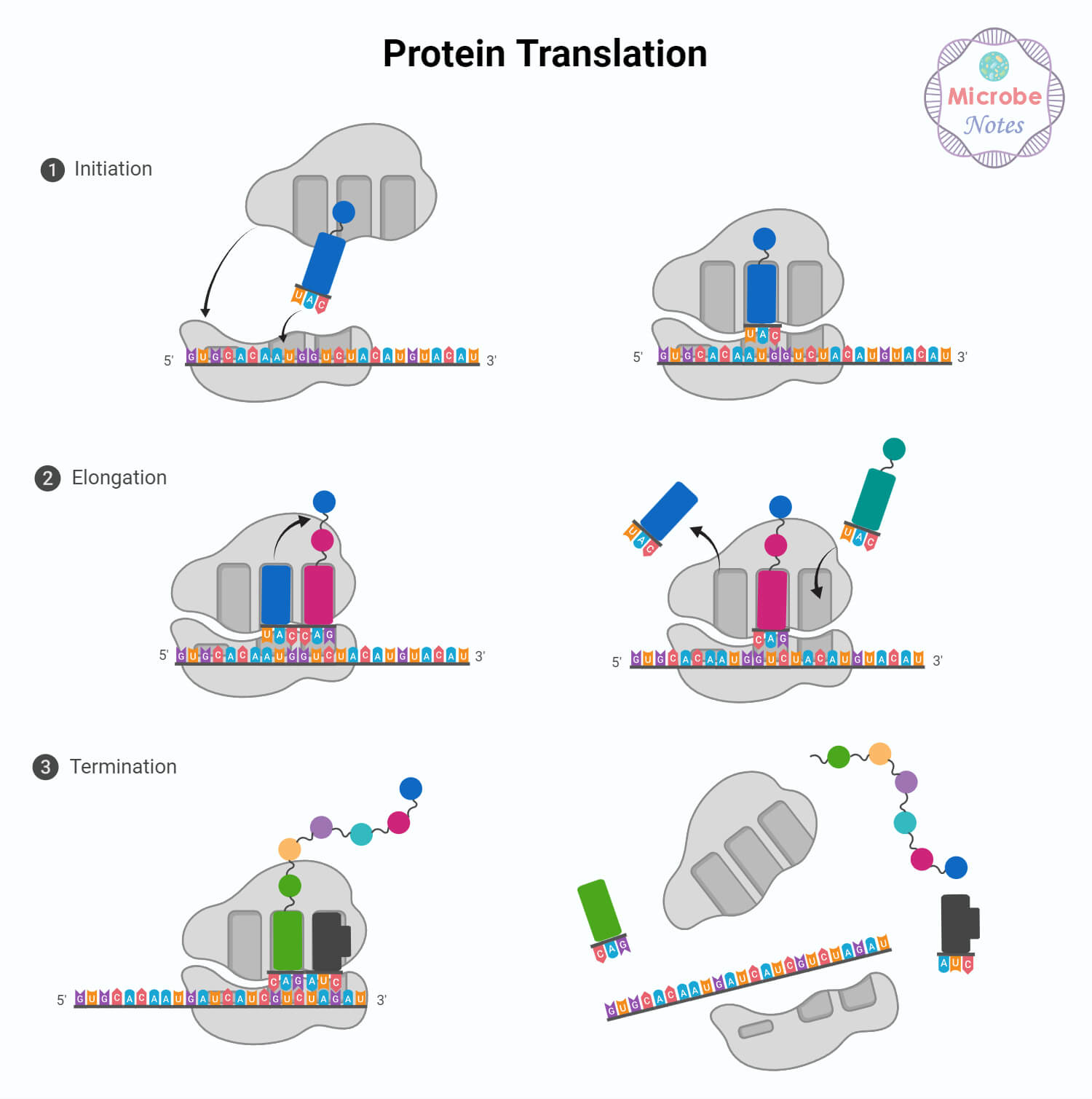
Translation Elongation
- The elongation of protein synthesis is aided by three protein factors i.e EF-Tu, EF-Ts, and EF-G.
- The ribosomal function is known to shift one codon at a time, catalyzing the processes that take place in its three sites.
- For every step, a charged tRNA enters the ribosomal complex and inserts the polypeptides that become one amino acid longer, while an uncharged tRNA departs. In prokaryotes, an amino acid is added at least every 0.05 seconds, which means that about 200 polypeptide amino acids are translated in 10 seconds.
- The bond created between each amino acid is derived from the Guanosine Triphosphate (GTP), which is similar to Adenosine Triphosphate (ATP).
- The three sites (A, P, E) all participate in the translation process, and the ribosome itself interacts with all the RNA types involved in translation.
- Therefore, three distinct steps are involved in translation, and these are;
- The mediation of elongation Factor-Tu (EF-Tu) in the entry of amino-acyl-tRNAs to the A site. This entails the binding of EF-Tu to GTP, which activates the EF-Tu-GTP complex to bind to tRNA. The GTP then hydrolyses to GDP releasing an energy-giving phosphate molecule, thus driving the binding of aminoacyl-tRNA to the A site. At this point the EF-Tu is released, leaving the tRNA in the A-site.
- Elongation factor EF-Ts then mediates the releasing of EF-Tu-GDP complex from the ribosomes and the formation of the EF-Tu-GTP.
- During this translocation process, the polypeptide chain on the peptidyl-tRNA is transferred to the aminoacyl-tRNA on the A-site during a reaction that is catalyzed by a peptidyl transferase. The ribosomes then move one codon further along the mRNA in the 5′ to 3′ direction mediated by the elongation factor EF-G. This step draws its energy from the splitting of GTP to GDP. Uncharged tRNA is released from the P-site, transferring newly formed peptidyl-tRNA from the A-site to the P-site.
Translation Termination
- Termination of the translation process is triggered by an encounter of any of the three stop codons (UAA, UAG, UGA). These triplet stop codons, however, are not recognized by the tRNA but by protein factors known as the release factors, (RF1 and RF2) found in the ribosomes.
- The RF1 recognizes the triplet UAA and UAG while RF2 recognizes UAA and UGA. A third factor also assists in catalyzing the termination process and it’s known as Release factor 3 (RF3).
- When the peptidyl-tRNA from the elongation step arrives at the P site, the release factor of the stop codon binds to the A site. These releases the polypeptide from the P site allowing the ribosomes to dissociate into two subunits by the energy derived from GTP, leaving the mRNA.
- After many ribosomes have completed the translation process, the mRNA is degraded allowing its nucleotides to be reused in other transcription reactions.
Protein Synthesis Video Animation (Amoeba Sisters)
Eukaryotes Protein Synthesis vs. Prokaryotes Protein Synthesis
| S.N. | Eukaryotes | Prokaryotes |
| 1. | The mRNA for translation is monocistronic, coding for a single gene of polypeptides | The mRNA for translation is polycistronic, thus coding for several genes of polypeptides |
| 2. | The three types of RNA polymerase are used for the synthesis of cellular RNA. | A single type of RNA polymerase is used to control the synthesis of the types of RNA molecules |
| 3. | It involves both subunits of the ribosomes i. e 40S and 60S subunits. | It involves 70S ribosomes |
| 4. | Transcription and translation take place separately hence they do not overlap. | Transcription and translation can overlap |
| 5. | The pre mRNA or an mRNA undergoes modification before they are translated. | The mRNA doesn’t undergo any modification before translation. |
| 6. | They do have a special initiator complex of tRNA. | A special initiator tRNA Met-tRNAf or Met – tRNA is used. |
| 7. | The starting amino acid is methionine. | The starting amino acid is N-formyl methionine |
| 8. | They have a single initiation and termination site. | They have several initiation and termination sites. |
| 9. | The Ribosomal Binding Site is Kozak sequence that is centered around the start codon | The ribosomal binding site (RBS) on mRNA is the Shine-Dalgarno sequence that lies -10 nucleotides ahead of the initiation codon. |
| 10. | Several initiation factors are involved in initiating the synthesise of the polypetide chain i.e eIF-2, (eIF-2, eIF-2al, eIF-a2, eIF-a | It involves three initiation factors IF-1, IF-2, and IF-3. |
| 11. | There are two chain elongation factors, EF-1 and EF-2 | There are three chain elongation factors, EF-Tu, EF-Ts, and EP-G. |
| 12. | There is a single release factor eRF for recognition of three termination codons (UAA, UAG, and UGA). | There are three release factors (RF-1 or RF-2 and RF-3) for recognition of termination codons. |
| 13. | The genetic code may differ in mitochondria and chloroplast. | The genetic code is the same in every prokaryotic organism. |
Protein Synthesis Inhibitors
Antimicrobial agents are used as protein synthesis inhibitors which include:
- Puromycin
- This is an antibiotic that is an analog of the terminal aminoacyl-adenosine part of aminoacyl-tRNA. This antibiotic inhibits protein synthesis by releasing prokaryotic polypeptides chains before they are completely synthesized. Its mechanism is achieved by joining its amino group to the carbonyl group of the growing polypeptide chain on the A-site forming an adduct that dissociates from the ribosome.
- Puromycin also contains an α-amino group similar to that on the aminoacyl-tRNA, which forms a covalently bound peptide bond with the carboxyl group of the growing peptide with puromycin residues, thus contributing to the dissociation of the ribosomes.
- Streptomycin
- This is a trisaccharide that has an effect on the binding activity of formyl methionyl-tRNA to ribosomes. This prevents the correct initiation of protein synthesis.
- Aminoglycoside antibiotics such as neomycin, kanamycin, and gentamycin which interfere with the decoding site in the 16s rRNA of the small subunit.
- Chloramphenicol inhibits the activity of peptidyl transferase.
- Erythromycin blocks translocation by binding to the 50S subunit
- Cycloheximide is used to block peptidyl transferase in eukaryotic ribosomes and it is used as a laboratory tool for blocking protein synthesis in eukaryotic cells.
- Diphtheria toxin has an A fragment that catalyzes the transfer of a single side chain of EF2 which blocks the translocation of the growing polypeptide chain.
References and Sources
- Microbiology by Prescott, 5th Edition
- 2% – https://www.biologydiscussion.com/cell/prokaryotes/comparison-of-synthesis-in-prokaryotes-and-eukaryotes/15520
- 1% – https://www.ncbi.nlm.nih.gov/books/NBK21424/
- 1% – https://www.cell.com/cell/pdf/0092-8674(89)90433-9.pdf
- 1% – https://en.wikipedia.org/wiki/Peptidyl_transferase
- 1% – https://bio.libretexts.org/LibreTexts/University_of_California_Davis/BIS_2A%3A_Introductory_Biology_(Facciotti)/Readings/SS1_2018_Lecture_Readings/SS1_2018_Lecture_11
- 1% – https://bio.libretexts.org/Courses/University_of_California_Davis/BIS_2A_(2018)%3A_Introductory_Biology_(Singer)/MASTER_RESOURCES/Translation%E2%80%94Protein_Synthesis*%23
- 1% – https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Boundless)/4%3A_Cell_Structure_of_Bacteria%2C_Archaea%2C_and_Eukaryotes/4.6%3A_Specialized_Internal_Structures_of_Prokaryotes/4.6A%3A_Ribosomes
- 1% – http://globalhealthprimer.emory.edu/targets-technologies/protein-synthesis.html
- <1% – https://www.thoughtco.com/protein-synthesis-translation-373400
- <1% – https://www.sciencedirect.com/topics/neuroscience/chloramphenicol
- <1% – https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/ribosomes
- <1% – https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/diphtheria-toxin
- <1% – https://www.sciencedaily.com/terms/peptide_bond.htm
- <1% – https://www.researchgate.net/publication/5558253_Inhibition_of_Helicobacter_pylori_Aminoacyl-tRNA_Amidotransferase_by_Puromycin_Analogues
- <1% – https://www.researchgate.net/publication/11483240_Analysis_of_tryptophanase_operon_expression_in_vitro_-_Accumulation_of_TnaC-peptidyl-tRNA_in_a_release_factor_2-depleted_S-30_extract_prevents_Rho_factor_action_simulating_induction
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4749135/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4307249/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3919579/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3323968/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2919715/
- <1% – https://www.nature.com/articles/s41389-018-0044-8
- <1% – https://www.cram.com/flashcards/mcat-biology-flashcards-1253204
- <1% – https://www.biologydiscussion.com/rna/rna-ribonucleic-acid-chemical-nature-types-and-rna-world/15450
- <1% – https://www.answers.com/Q/What_is_the_anticodon_for_methionine
- <1% – https://www.annualreviews.org/doi/full/10.1146/annurev.biochem.72.110601.135450
- <1% – https://wikimili.com/en/Peptidyl_transferase
- <1% – https://teaching.ncl.ac.uk/bms/wiki/index.php/Translation
- <1% – https://quizlet.com/77160525/genetics-unit-iv-ch-12-from-p322-on-ch-13-and-ch-15-flash-cards/
- <1% – https://quizlet.com/477610163/biochemistry-ch-7-flash-cards/
- <1% – https://quizlet.com/291704106/genetics-chapter-13-usf-flash-cards/
- <1% – https://quizlet.com/20669105/chapter-13-translation-flash-cards/
- <1% – https://pediaa.com/difference-between-mrna-and-trna-and-rrna/
- <1% – https://microbenotes.com/rna-polymerase/
- <1% – https://flexbooks.ck12.org/cbook/ck-12-biology-flexbook-2.0/section/4.7/primary/lesson/translation-of-rna-to-protein-bio/
- <1% – https://febs.onlinelibrary.wiley.com/doi/pdf/10.1111/j.1432-1033.1983.tb07643.x
- <1% – https://en.wikipedia.org/wiki/Peptidyl-tRNA
- <1% – https://en.wikipedia.org/wiki/MRNA
- <1% – https://en.wikipedia.org/wiki/EF-Tu
- <1% – https://ecampusontario.pressbooks.pub/microbio/chapter/protein-synthesis-translation/
- <1% – https://barbadosunderground.files.wordpress.com/2015/03/antimicrobial-drugs-a-fading-miracle.pdf
- <1% – https://answers.yahoo.com/question/index?qid=20070613182552AAYieJn
- <1% – http://thebiologyprimer.com/transcription-rna-processing-and-translation


Thank you for such valuable notes