RNA splicing is a form of RNA processing in which a newly made precursor messenger RNA (mRNA) is transformed into a mature RNA by removing the non-coding sequences termed introns.
- The process of RNA splicing involves the removal of non-coding sequences or introns and joining of the coding sequences or exons.
- RNA splicing takes place during or immediately after transcription within the nucleus in the case of nucleus-encoded genes.
- In eukaryotic cells, RNA splicing is crucial as it ensures that an immature RNA molecule is converted into a mature molecule that can then be translated into proteins. The post-transcriptional modification is not necessary for prokaryotic cells.
- RNA splicing is a controlled process that is regulated by various ribonucleoproteins.
Interesting Science Videos
What are Introns?
Introns are non-coding DNA sequences present within a gene that are removed by the process of RNA splicing during maturation of the RNA transcript.
- The word ‘introns’ is used to denote both the DNA sequences within the gene and the corresponding sequence in RNA transcripts.
- Introns are common in the protein-coding nuclear genes of most jawed invertebrates other eukaryotic organisms along with unicellular organisms like bacteria.
- Similarly, the mitochondrial genomes of jawed vertebrates are almost entirely devoid of introns whereas those in other eukaryotes have many introns.
- During RNA splicing, the introns between the exons are removed to connect two different exons that then code for messenger RNA.
- Introns are crucial because the variation in the protein bio-product formed is greatly enhanced by alternative splicing in which introns take part in prominent roles.
- Introns have a donor site (5′ end), a branch site (near the 3′ end), and an acceptor site (3′ end) that are required for splicing.
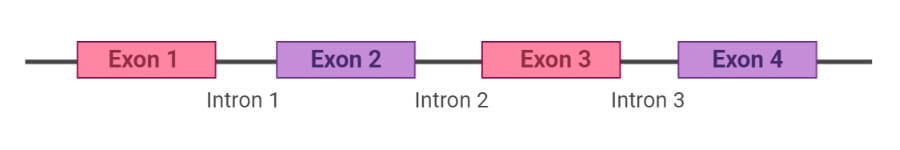
What are Exons?
Exons are protein-coding DNA sequences that contain the necessary codons or genetic information essential for protein synthesis.
- The word ‘exon’ represents the expressed region present in the genome.
- The exosome is the term used to indicate the entire set of all exons present in the genome of the organisms.
- In genes coding for proteins, exons include both the protein-coding sequence and the 5’ and 3’ untranslated regions.
- Exons are found in all organisms ranging from jawed vertebrates to yeasts, bacteria, and even viruses.
- In the human genome, exons account for only 1% of the total genome while the rest is occupied by intergenic DNA and introns
- Exons are essential units in protein synthesis as they carry regions composed of codons that code for various proteins.
- Alternative splicing enables exons to be arranged in different combinations, where different configuration results in different proteins.
- A process similar to alternative splicing is exon shuffling where exons or sister chromosomes are exchanged during recombination.
What is Spliceosome?
A spliceosome is a large and complex molecule formed of RNAs and proteins that regulate the process of RNA splicing.
- The spliceosome is composed of five small nuclear RNAs (snRNA) and about 80 protein molecules.
- The combination of RNAs with these proteins results in the formation of an RNA-protein complex termed as small nuclear ribonucleoproteins (snRNPs).
- These are mostly confined within the nucleus where they remain associated with the immature pre-RNA transcripts.
- These spliceosomes, in addition to working on RNA-RNA interactions, are also involved in RNA-protein interactions.
- The spliceosome functions as an editor that selectively cuts out unnecessary and incorrect materials (introns) to produce a functional final-cut.
- All spliceosomes are involved in both the removal of introns and the ligation of remaining exons.
- Another set of spliceosomes termed ‘minor spliceosomes’ are also found in eukaryotic cells which have less abundant RNAs and are involved in the splicing of a rare class of pre-mRNA introns.
RNA Splicing Process/ Mechanism
- The process of RNA splicing begins with the binding of the ribonucleoproteins or spliceosomes to the introns present on the splice site.
- The binding of the spliceosome results in a biochemical process called transesterification between RNA nucleotides.
- During this reaction, the 3’OH group of a specific nucleotide on the intron, which is defined during spliceosome assembly, causes a nucleophilic attack on the first nucleotide of the intron at the 5’ splice site.
- This causes the folding of the 5’ and 3’ ends, resulting in a loop. Meanwhile, the adjacent exons are also brought together.
- Finally, the looped intron is detached from the sequence by the spliceosomes.
- Now, a second transesterification reaction occurs during the ligation of adjacent exon segments.
- In this case, the 3’OH group of the released 5’ exon then performs an electrophilic attack on the first nucleotide present just behind the last nucleotide of the intron at the 3’ splice site.
- This causes the binding of the two exon segments along with the removal of the intron segment.
- Earlier, the intron released during splicing is thought of as a junk unit. Still, it has been recently observed that these introns are involved in other processes related to proteins after their removal.
- Besides the spliceosomes, another group of protein/ enzymes termed ‘ribozymes’ are also involved in the control and regulation of the splicing process.
Types of RNA Splicing
1. Self-splicing
- Self-splicing is a type of RNA splicing which occurs in some rare introns that are capable of promoting phosphodiester bond cleavage and formation without the help of other proteins or spliceosomes.
- These introns are unique as they can mediate their excision from precursor RNA and the subsequent ligation of the flanking exons in a simple salt buffer.
- This self-splicing reaction is facilitated by the tertiary structure of the intron, which provides the ability to recognize the splice sites of the precursor RNA and to perform the cutting and ligation reactions in a very precise manner.
- The sequence present in such introns performs as a ribozyme that regulates the overall process.
- There are three types of self-splicing introns that are grouped as Group I, Group II, and Group III.
- Group I and Group II introns perform the splicing process in a mechanism similar to that by spliceosomes. These suggest that these introns might be evolutionarily related to the spliceosomes.
- During self-splicing, the 5′ splice site is recognized by a short sequence element in the intron called the internal guide sequence.
- Besides, other strongly conserved sequences of the introns called P, Q, R, and S are needed to ‘catalyze’ the cutting and ligation reactions.
- Self-splicing follows a similar mechanism involving two transesterification reactions resulting in the removal of introns and ligation of exons.
2. Alternative Splicing
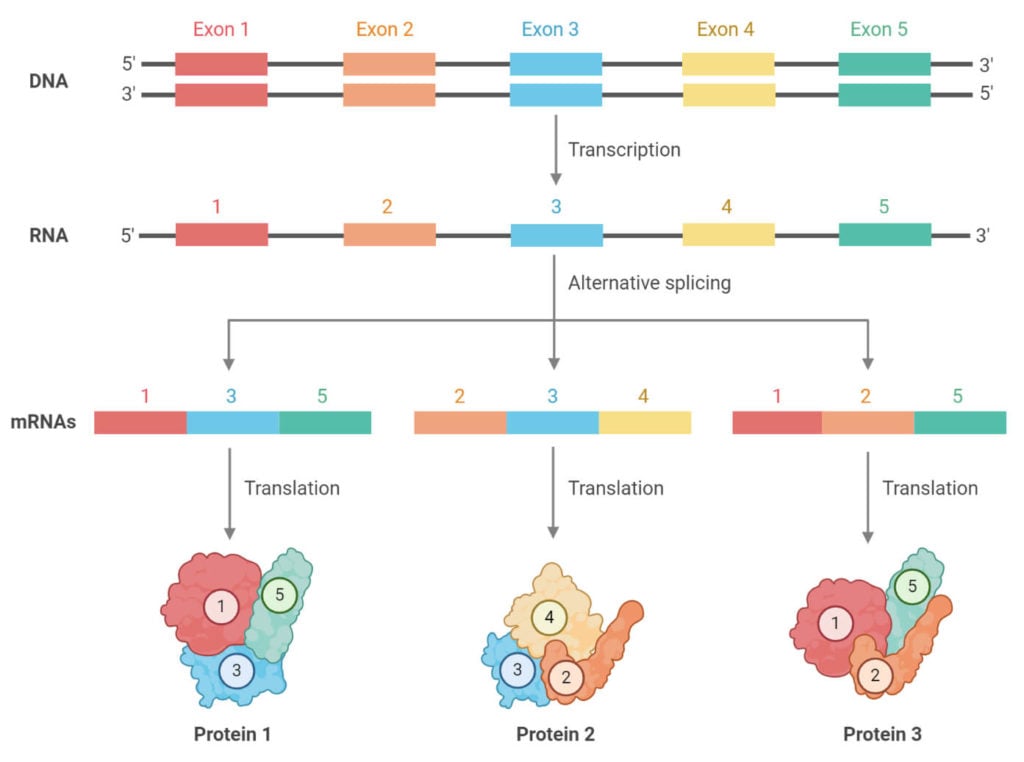
- Alternative splicing is a splicing process resulting in a varying composition of exons in the same RNA and creating a range of unique proteins.
- Alternative splicing of pre-mRNA is an essential mechanism to enhance the complexity of gene expression, and it also plays a vital role in cellular differentiation and organism development.
- Alternative splicing enables exons to be arranged in different combinations where different configuration results in different proteins
- The process of alternative splicing might occur either by skipping or extending some exons or by retaining particular introns, resulting in different varieties of mRNA formed.
- Regulation of alternative splicing is a complex process in which numerous components interact with each other, including cis-acting elements and trans-acting factors.
- The process is further guided by the functional coupling between transcription and splicing.
- Additional molecular features, such as chromatin structure, RNA structure, and alternative transcription initiation or alternative transcription termination, collaborate with these basic components to generate the protein diversity due to alternative splicing.
- Alternative splicing is also essential for other functions like the identification of novel diagnostic and prognostic biomarkers, as well as new strategies for therapy in cancer patients.
- Thus, alternative splicing has a role in almost every aspect of protein function, including binding between proteins and ligands, nucleic acids or membranes, localization, and enzymatic properties.
3. tRNA splicing
- Like in mRNA, the genes in tRNA are also interrupted by introns, but here the splicing mechanism is quite different.
- Splicing in tRNA is catalyzed by three enzymes with an intrinsic requirement for ATP hydrolysis.
- The process of tRNA splicing occurs in all three major lines of descent, the Bacteria, the Archaea, and the Eukarya, but the mechanism might differ in bacteria and higher organisms.
- In bacteria, the introns in the tRNA are self-splicing.
- In Archaea and Eukarya, however, the tRNA splicing reaction occurs in three steps where each step is catalyzed by a distinct enzyme, each of which can function interchangeably on all of the substrates.
- In the first step, the pre-tRNA is cleaved at the two splice sites by an endonuclease, resulting in two tRNA half molecules and a linear intron with 5’-OH and 3’-cyclic PO4 ends.
- The cleavage is then followed by the ligation of the two RNA half molecules in the presence of a tRNA ligase enzyme.
- Finally, the PO4 ends produced from splicing are transferred to NAD in a process catalyzed by nicotinamide adenine dinucleotide (NAD)-dependent phosphotransferase.
RNA Splicing and Spliceosome Video Animation By Frank Lectures
RNA Splicing errors
- The splicing of nuclear pre-mRNAs is a fundamental process required for the expression of most metazoan genes. However, errors in splicing might occur due to mutations that result in various splicing-related diseases.
- Mostly in alternative splicing, an erroneous splicing result in biological products that are not functional.
- Errors during splicing might occur due to mutations at the splice site, which causes loss of exons or inclusion of an intron disrupting the function of the RNA sequence.
- Similarly, displacement of a splice site might also cause the formation of longer or shorter exons, resulting in erroneous products.
- In living organisms like plants, stress-induced alternative splicing associated with various metabolic pathways might bring changes in the normal functioning of the plant.
- The chances of erroneous splicing is more in eukaryotic cells with high levels of alternative splicing containing splice sites that have evolved to offer a weak binding potential for components of the spliceosome
- The accuracy of splice-site pairing is also limited by the accuracy of transcription as the transcription machinery makes a mistake once in every 103–105 nucleotide insertion step.
RNA Splicing Application
There are various biological, medical applications associated with pre-mature RNA splicing, some of which are:
- Pre-mRNA splicing is a fundamental process in cellular metabolism that plays an essential role in generating protein diversity. The diversity is brought about by changes in the number and sequence of exons and introns present in the RNA sequence.
- RNA splicing also helps in the regulation of gene and protein content in the cell.
- Splicing of RNA sequences assists the process of evolution of new and improved proteins.
- Various aberrant splicing isoforms act as markers for cancer and as targets for cancer therapy.
- Pre-mRNA splicing is a key to the pathology of cancers where it regulates the three functional aspects of cancer: proliferation, metastasis, and apoptosis.
References
- Verma PS and Agarwal VK (3005). Cell Biology, Genetics, Molecular Biology, Evolution and Ecology. Multicoloured Edition
- Hsu, S. N., & Hertel, K. J. (2009). Spliceosomes walk the line: splicing errors and their impact on cellular function. RNA biology, 6(5), 526–530. https://doi.org/10.4161/rna.6.5.9860
- Wang, Y., Liu, J., Huang, B. O., Xu, Y. M., Li, J., Huang, L. F., Lin, J., Zhang, J., Min, Q. H., Yang, W. M., & Wang, X. Z. (2015). Mechanism of alternative splicing and its regulation. Biomedical reports, 3(2), 152–158. https://doi.org/10.3892/br.2014.407
- Di, C., Syafrizayanti, Zhang, Q. et al.Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ 26, 1181–1194 (2019). https://doi.org/10.1038/s41418-018-0231-3
- John Abelson, Christopher R. Trotta, and Hong Li (1998). tRNA Splicing. THE JOURNAL OF BIOLOGICAL CHEMISTRY. Vol. 273, No. 21, Issue of May 22, pp. 12685–12688, 1998
- Winter, A. J., Groot Koerkamp, M. J., & Tabak, H. F. (1990). The mechanism of group I self-splicing: an internal guide sequence can be provided in trans. The EMBO journal, 9(6), 1923–1928.
- https://www.khanacademy.org/science/biology/gene-expression-central-dogma/transcription-of-dna-into-rna/a/eukaryotic-pre-mrna-processing


This is very excellent
I want know about the pre-mRNA splicing in detail