Interesting Science Videos
What is RNA?
Its full form is Ribonucleic acid. This is a polymer of subunits joined by phosphodiester bonds. It is a single-stranded nucleic acid similar to DNA but having ribose sugar rather than deoxyribose sugar and uracil instead of thymine as one of the nucleotide bases.
Types of RNA
RNA polymerase synthesizes RNA from DNA that is functionally for protein-coding (messenger RNA, mRNA) or non-coding (RNA genes). Because of these functions, RNA molecules are of following types:
- messenger RNA (mRNA) – It is the RNA that carries information from DNA to the ribosomes (site of protein synthesis) in the cell. The mRNA code sequences determine the amino acid sequence in the protein that is produced.
- ribosomal RNA (rRNA) – It incorporates into the ribosomes.
- transfer RNA (tRNA) – It is used to transfer specific amino acids to growing polypeptide chains at the ribosomal site of protein synthesis during translation.
- small nuclear RNA (snRNA)
- microRNA (miRNA) – They are used to regulate gene activity; They are tiny (~22 nucleotides) RNA molecules that regulate the expression of messenger RNA (mRNA) molecules.
- small nucleolar RNA (snoRNA)
- long non-coding RNA (lncRNA)
- catalytic RNA (ribozymes) which functions as an enzymatically active RNA molecule.
Read Also: RNA- Properties, Structure, Types, and Functions
1. Messenger RNA (mRNA): Structure and Functions
- It is synthesized in the cell nucleus and then transported out of the cell to facilitate protein synthesis and code sequencing on proteins.
- The mRNA is translated into polypeptides.
- It comes in a wide range of sizes which reflects the polypeptide size it encodes.
- Most cells produce thousands of different mRNA molecules in small amounts, which are translated into peptides that are needed for use by the cell.
- Many mRNA molecules are common to most cells which encode for proteins that guard cell metabolisms such as the enzymes used In glycolysis.
- Some types of mRNA are specific for certain types of cells, which encode for the proteins that are needed for the function of that particular cell such as mRNA for hemoglobin is found in Red Blood Cells (RBCs).
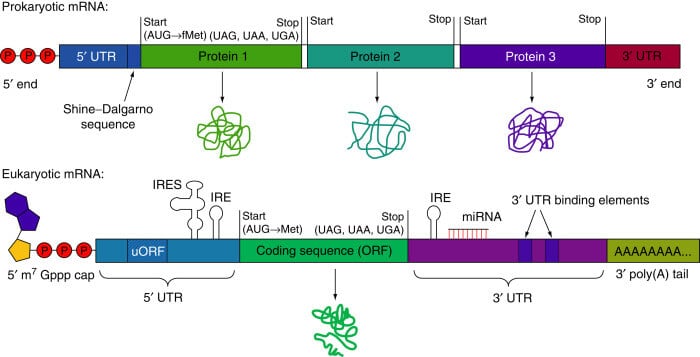
Figure: Prokaryotic and Eukaryotic Messenger RNA (mRNA). Image Source: ScienceDirect.
Structure and Functions of mRNA
A mature mRNA of the eukaryotic cell is made up of 5 subunits which are:
- 5′ Cap
- This is an altered nucleotide on the 5′ end of the primary transcript on mRNA, by a process known as mRNA capping. mRNA capping plays a primary role in the regulation and creation of mature mRNA in protein synthesis (translation). However, note that mitochondrial mRNA and chloroplastic mRNA are not capped.
- The 5′ end has a guanine nucleotide that is connected to mRNA by a 5′ to 5′ triphosphate linkage. The guanine nucleotide is methylated by methyltransferase on the 7th position, referring to it as a 7-methyl guanylate cap (m7G) after capping.
- This cap is chemically similar to the 3′ end of the RNA molecule.
- This cap is uniquely found in small nuclear RNAs (snRNA) with a 5′ trimethylguanosine caps and in the long non-coding RNA (lncRNA) as 5′ monomethyl phosphate cap.
- In bacteria and some organisms, the mRNA is capped with NAD+, NADH, or 3′ dephospho-coenzyme A.
- In all organisms, mRNA molecules are decapped by a mechanism known as messenger RNA decapping.
- 5′ untranslated region (5′ UTR)
- This is the region of mRNA that is directly upstream from the initiation codon.
- It is essential for the regulation of translation of a transcript in viruses, prokaryotes, and eukaryotes.
- Some part of the 5′ untranslated region is sometimes translated into protein products, which then regulates the translation of the main coding sequence on the mRNA.
- However, in some organisms, the 5′ UTR is not translated therefore it forms the complex secondary structure to regulate translation.
- Coding region
- This is the region on RNA that codes for proteins
- The coding region in DNA flanks the promoter sequence which on proceeding to transcription, the RNA polymerase binds to the promoter sequence moving along the template strand to the coding region. the RNA polymerase adds RNA nucleotides that are complementary to the coding region forming the mRNA, by substituting thymine with uracil. The mechanism takes place until the occurrence of the termination sequence.
- Three prime untranslated region (3′ UTR)
- This is the part of mRNA that follows the translation termination codon. This unit contains the regulatory regions that post-transcriptionally influence gene expression.
- The 3′ UTR is not translated to proteins, same as the 5′ untranslated region.
- However, regulatory regions found in the 3 UTR influences polyadenylation, the efficiency of translation, translation localization, and the stability of mRNA.
- It has binding sites that are used for regulatory proteins and microRNAs (miRNAs), which regulate and decreases gene expression of various mRNAs by either inhibiting translation or directly causing transcript degradation.
- The 3′ UTR also has a silencer region that binds to the repressor proteins, which inhibits the expression of the mRNA.
- Several 3′ UTRs also contain Adenine-Uracil (AU) rich elements sequences as (AAUAAA) which directs adding adenine residues known as poly (A) tail to the end of the mRNA transcript.
- The 3′ UTR also contains sequences that promote proteins association with mRNA with the cell cytoskeleton.
- It transports it to or from the cell nucleus or performs other types of cell localization.
- Generally, the 3′ UTR helps to regulate genes and ensure the right genes are expressed correctly at the right time.
- Poly A tail
- The poly(A) tail is made up of multiple adenosine monophosphates i.e it is a stretch of RNA that is made up of adenine bases.
- It is bound with poly-A binding proteins (PABP) and they play a primary role in the regulation of mRNA translation, stability, and export.
- The poly (A) tail that is bound to the PABP interacts with proteins associated with the 5′ end of the transcript, which causes the circulation of the mRNA that promotes the protein translation process.
- The addition of poly-A tail to mRNA is known as polyadenylation.
- Eukaryotic polyadenylation helps produce mature messenger RNA (mRNA) used in the translation.
- In several bacteria, the poly-A tail has been useful in promoting the degradation of mRNA, meaning that, poly-A forms the larger process of gene expression.
- The role of poly-A in polyadenylation begins as the transcription of the genes terminates.
- The poly-A tail is essential for nuclear export, translation, and stability of mRNA.
- As the tail shortens with time and becomes short enough, the synthesis of mRNA is degraded enzymatically.
2. Ribosomal RNA (rRNA): Structure and Functions
Structure
- Ribosomal ribonucleic acid (rRNA) is the type of RNA that is part of the ribosomes.
- It is defined as the molecular machine that catalyzes the synthesis of proteins.
- It makes up to 60% of the weight of the ribosomes since they play a major part in the functions of the ribosomes such as binding to mRNA and the recruitment of tRNA, and the catalyzation of peptide bond formation between amino acids.
- They also determine the shape of ribosomes using the rRNA core.
- rRNA has a distinctive three-dimensional shape which is made up of internal loops and helices that creates specific sites, A, P, and E, within the ribosome.
- The P site functions as a binding site for the growing polypeptide, the A site acts as an anchor to an incoming tRNA charged with an amino acid.
- After the formation of the peptide bond, the tRNA binds to the E site, briefly before leaving the ribosomes. After peptide bond formation, the tRNA binds briefly to the E site before leaving the ribosome.
- Additionally, the rRNA has a site for binding to the ribosomal proteins, and it analyses and distinguishes the residues of RNA from proteins.
- The proteins on the ribosome surface stabilize its structure by interacting with the rRNA core.
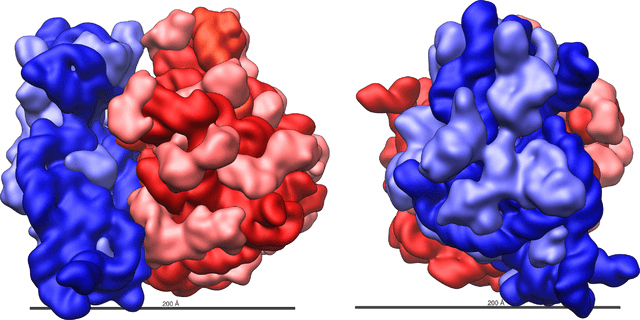
Figure: Three-dimensional views of the ribosome, showing rRNA in dark blue (small subunit) and dark red (large subunit). Lighter colors represent ribosomal proteins. Source: Vossman (Wikipedia).
Function
- The rRNA is synthesized or transcribed in the cell nucleus, specifically in the nucleoli. The nucleoli play a major role in the biogenesis of ribosomes via the sequestration of ribosomal proteins.
- Both prokaryotic and eukaryotic ribosomes are made of a larger and smaller subunit and these two units come together during the translation of mRNA.
- The small subunits of the prokaryotes are made of an RNA molecule of about 1500 nucleotides in length with a Svedberg coefficient of 16S.
- The small subunit along with ribosomal proteins has a sedimentation rate of the 30S.
- This is paired with the larger subunit, having two RNA molecules – one that is nearly 3000 nucleotides (23S) in length and the other is a short sequence of 120 nucleotides (5S). These RNA molecules are accompanied by proteins that give rise to the larger 50S subunit.
- The eukaryotic ribosomes are made up of two subunits also i.e the large subunit 60S and the small subunit 40S.
- The small subunit is made up of two short rRNA molecules that are less than 200 nucleotides in length (5S and 5.8S), and the large subunit which is made up of two large molecules that are longer, one which has over 5kb (28S) and a second one with 2kilobases (18S).
- The 28S, 18S, and 5.8S molecules are produced by the processing of a single primary transcript from a cluster of identical copies of a single gene. The 5S molecules are produced from a different cluster of identical genes.
- Totally, the eukaryotic ribosome has a Svedberg coefficient of the 80S.
- Eukaryotic cells also have rRNA in mitochondria and chloroplast.
- Ribosomes are associated with the endoplasmic reticulum or can be free-floating in the cell cytoplasm.
3. Transfer RNA (tRNA): Structure and Functions
- This is the non-coding RNA molecule that carries amino acids to the ribosomes, from the growing peptide chain (mRNA nucleotide sequence. Therefore, the tRNA acts as the intermediate between nucleotide and amino acid sequences.
- They are ribonucleotides, therefore, they form a hydrogen bond with mRNA, and form ester links with amino acids which combine the mRNA and amino acids during translation.
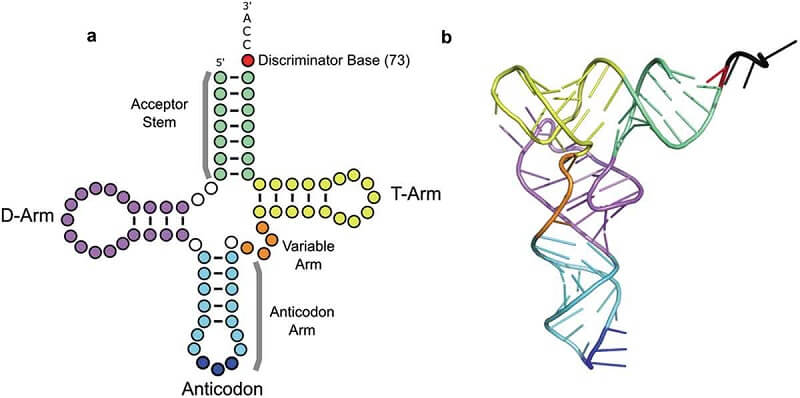
Figure: Transfer RNA (tRNA) – (a) tRNAs are represented as cloverleaf structures in two dimensions. (b) In three dimensions, tRNAs fold into an L-shape stabilized by intramolecular base-pairing shown here by the tRNA Phe structure [PDB: 1HEZ; [20]]. In both diagrams, the tRNA structural elements are colored: acceptor stem (green), dihydrouridine (D)-arm (purple), anticodon stem (light blue), anticodon (bases 34, 35, 36 in dark blue), variable arm (orange), T-arm (yellow) and the discriminator base (red). Image Source: https://doi.org/10.1080/15476286.2019.1646079
Structure and Function
- It is a small RNA chain of about 80 nucleotides.
- During translation, tRNA transfers specific amino acids corresponding to the mRNA sequence to the growing polypeptide chain in the ribosome.
- tRNA pairs with mRNA complementarity in a parallel manner with each of its base pairs having three nucleotides paired to mRNA.
- tRNAs are coded by short molecules of 70-90 nucleotides (5nm).
- The set of the three nucleotides on the mRNA is known as a codon, while the corresponding sequence on tRNA is known as an anticodon.
- The base pairing of the codon and the anticodon forms a translation mechanism
- At the end of the tRNA 3′ hydroxyl base, there is an anticodon amino acid sequence that is attached, linking the ribosomes to form a peptide bond, thus elongating the polypeptide chain.
- Therefore, tRNA major parts are the anticodon and the 3′ hydroxyl group terminal.
- Other parts of the tRNA structure are the D-arm and the T-arm, which are highly specific and are highly effective.
- tRNAs have a sugar-phosphate backbone which gives it directionality.
- One end of the tRNA has a reactive phosphate group, which is attached to the fifth carbon atom of the ribose (5′) and another end which has a free hydroxyl group on the third carbon (3′), giving rise to the 5′ to 3′ ends of RNA.
- The 3′ terminal end has three bases CCA (Cytosine, cytosine, adenine) which make part of the acceptor arm of the molecule which is covalently attached to the hydroxyl group on the ribose sugar.
- The acceptor arm also contains parts of the 5’ end of the tRNA, made up of 7-9 nucleotides on the opposite ends of the molecule base pairing with each other.
- The anticodon loop which is recognized by the aminoacyl tRNA synthetase (AATS) is paired to mRNA and it determines the amino acid that attaches to the acceptor’s arm.
- The AATS reads and recognizes the D-arm from the 5′ end of tRNA.
- The D-arm plays a major role in stabilizing the structure of RNA; it affects and influences the kinetics and accuracy of translation at the ribosomes.
- The T-arm also influences the tRNA effect on the translation by interaction with the ribosomes.
- The D-arm, T-arm, and the anticodon loop combined resembles a cloverleaf. When RNA folds into a tertiary structure, it becomes L-shaped with an extended structure of the acceptor stem, T-arm, anticodon loop, and D-arm.
4. Small Nuclear RNA (snRNA): Structure and Functions
- During DNA transcription for mRNA, rRNA, and tRNA, primary transcripts are processed in the nucleus, so as to produce functional elements which are to be exported to the cytosol. The role of the small nuclear RNA (snRNA) is to mediate some of these processes.
- The snRNA are transcribed by RNA polymerase II or RNA polymerase III, of about 150 nucleotides.
- snRNA has different genes in multiple copies, which play different roles in the synthesis of other RNA classes, such as, snRNA which is part of the spliceosomes that help in the conversion of pre-messenger RNA (hnRNA) into mRNA by excising the introns and splicing the exons.
- They also mediate the regulation of transcription factors and RNA polymerase II.
- They also maintain telomeres.
- snRNA associate with specific proteins and complexes known as small nuclear ribonucleoproteins also termed as snRNPs.
5. Small Nucleolar RNA (snoRNA): Structure and Functions
- They are small RNAs of about 60-300 nucleotides found in the cell nucleolus that play different functions in the cell.
- They play a role in the synthesis of ribosomes, by cutting the large RNA precursor of the 28S, 18S, and 5.8S
- They chemically modify many of the nucleotides in rRNA, tRNA, and snRNA molecules by adding groups such as methyl groups to ribose.
- They also help in the splicing of pre-mRNA to different forms of mature mRNA.
- One type of snoRNA serves as a template for the synthesis of the telomeres.
- In the vertebrates, the snoRNAs are made from introns that removed during transcription.
6. MicroRNAs (miRNAs): Structure and Functions
- MicroRNA is a small non-coding RNA that is single-stranded, containing 22 nucleotides. Its size is estimated to be the same as that of siRNAs.
- It is found in plants, all animals, and some viruses, with its primary role in RNA silencing and post-transcriptional gene expression regulation.
- Humans generate about 1000miRNAs.
- These miRNAs are encoded in the genome by stand-alone genes or in portions of introns of the genes where they regulate the mRNA.
- They are expressed in some cell types at certain times during the differentiation of these cell types.
- Their role in gene regulation by regulating the expression of mRNA is achieved in two ways:
- By destroying the mRNA when the sequences are evenly matched especially in plants
- By repressing the translation of mRNA when the sequences are matched partially.
- These characteristic roles of miRNA are attributed to two of its features
- their small size makes it easy to rapidly transcribe from their gene
- they do not need a translated protein element for them to regulate mRNA gene expression for either partial or evenly matched gene sequences.
- Genomic studies of mammalian gene expression have revealed that one or more miRNAs are bound to an mRNA that has been transcribed from DNA.
- A single miRNA can bind to about 200 different mRNA targets, favored by the presence of multiple miRNA binding sites on mRNA.
- This allows coordinated mRNA translation.
7. Long Non-coding RNA (lncRNA): Structure and Functions
- This is a heterogeneous group of non-coding transcript RNA that are 200 nucleotides in size.
- They are the largest mammalian non-coding transcriptome.
- An estimated 8000lncRNAs are encoded in the human genome.
- Major functions of lncRNA are still unknowns, however, some scientific evidence indicates its role in gene regulation and physiological mechanism involvements.
- Some of its known functions in gene regulation mechanisms include:
- splicing
- translation
- imprinting
- transcription
- Inactivation of one of the two X chromosomes in female vertebrates by a type of lncRNA known as XIST-RNA
- They play a role in bringing the enhancer and promoter regions of genes to close together by looping, which helps in the regulation of gene transcription.
NOTE:
The non-coding RNAs (tRNA, rRNA, snoRNA, snRNA, miRNA, and lncRNA) account of three-quarters of transcription that takes place in the cell nucleus.
References and Sources
- Microbiology by Prescott
- https://www.futurelearn.com/courses/translational-research/0/steps/14201
- The Biology Dictionary-rRNA Notes
- The Biology Dictionary -tRNA Notes
- https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx
- http://www.phschool.com/science/biology_place/biocoach/transcription/difgns.html
- https://bio.libretexts.org/Bookshelves/Genetics/Book%3A_Working_with_Molecular_Genetics_(Hardison)/Unit_III%3A_The_Pathway_of_Gene_Expression/10%3A_Transcription%3A_RNA_polymerases
- https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Book%3A_Biology_(Kimball)/06%3A_Gene_Expression/6.02%3A_The_Transcription_of_DNA_into_RNA
- https://www.ebi.ac.uk/chebi/searchId.do?chebiId=74035
- https://www.genome.gov/genetics-glossary/Transfer-RNA
- http://bioscience.jbpub.com/cells/MBIO5245.aspx
- https://en.wikipedia.org/wiki/Small_nuclear_RNA#:~:text=Small%20nuclear%20RNA%20(snRNA)%20is,snRNA%20is%20approximately%20150%20nucleotides.
- https://www.futurelearn.com/courses/translational-research/0/steps/14201
- https://www.news-medical.net/life-sciences/-Types-of-RNA-mRNA-rRNA-and-tRNA.aspx
- https://www.britannica.com/science/RNA#ref340177
- https://en.wikipedia.org/wiki/Polyadenylation
- https://en.wikipedia.org/wiki/Three_prime_untranslated_region
- https://en.wikipedia.org/wiki/Coding_region
- https://en.wikipedia.org/wiki/Five_prime_untranslated_region
- https://en.wikipedia.org/wiki/Five-prime_cap
- https://en.wikipedia.org/wiki/Messenger_RNA
- https://www.genome.gov/genetics-glossary/messenger-rna#:~:text=Messenger%20RNA%20(mRNA)%20is%20a,cytoplasm%20where%20proteins%20are%20made.

Its a great job, very thanks!
So , interesting! Thanks
Thank you sir
Very good information. I want pdf file of that information please.