Chromatin is a relative term used to describe the association of DNA and histone proteins in the formation of chromosomes. It is observed that chromatin is made of proteins (50-60%), DNA (30-40%), and RNA (1-10%). The chromatin structure is referred to as ‘beads on a string’ because the DNA threads are wrapped around histone proteins and observed during the replication process (mitosis/meiosis).
Chromatin fibers are stainable; hence, it is easy to find the cell replication stages based on chromatin bands. Chromatin functions include packaging genetic material to fit inside the nucleus and regulation of gene expression via replication of DNA, transcription, chromosome segregation, and recombination. Based on the structure, compaction, and location in a chromosome, chromatin fibers are classified as heterochromatin, euchromatin, and centromeric chromatin.
- Euchromatin constitutes the chromatin structure associated with the active transcription of genes.
- Heterochromatin makes up less accessible chromatin fibers that are associated with silencing.
- Centromeric chromatin are fibers recruited for spindle binding during chromosomal segregation.
Interesting Science Videos
What is Euchromatin?
Euchromatin is chromatin fibers with transcriptionally active genes, with wider spaces between the nucleosomes (DNA + histone protein = repeating units of chromatin structure); hence they are also called Open Chromatin.
- Euchromatin fibers facilitate active transcription by allowing histone modifications and higher accessibility for transcription machinery to act.
- It is observed that euchromatin consists of more nucleosome beads with specific nucleosomal positioning.
- This exact position of nucleosome aids in the transcription regulation associated with precise recognition by histone proteins of DNA sequence motifs.
- According to the article ‘Finishing the euchromatic sequence of the human genome,’ 92% comprises the euchromatin structure.
Euchromatin Structural Composition
Euchromatin is made up of repetitive units of nucleosomes. Nucleosomes are roughly 11 nm in diameter and comprise DNA fibers wrapped around the histone protein. Euchromatin’s spacing between individual nucleosomes is wider, allowing transcriptional protein complexes to access the DNA motifs easily.
- DNA helices are condensed, allowing less than two helical turns to wrap around the histone proteins.
- Approximately 200 base pairs coil around the histones.
- The DNA that links two nucleosomes is known as the Linker DNA, containing around 0 to 80 base pairs.
- Histone protein core comprises four pairs of polypeptide chains, two pairs of each polypeptide – H2A, H2B, H3, and H4, the histone octamers.
- Histones possess a ‘tail structure’ that varies in the amino acid sequence, and via methylation and acetylation processes, it acts like control switches, swapping between different chromatin conformations.
- Methylation of the fourth lysine present in the histone tail induces euchromatin conformation. Hence it is also used as a marker during experiments to observe euchromatin structures.
Euchromatin Appearance
Euchromatin appears as ‘beads on a string’ by magnifying the chromosomal structures. In electron and optical microscopic settings, euchromatin fibers stain lighter than heterochromatin due to wider nucleosome spacing and more open structure. The widely used G-banding (Giemsa) staining technique differentiates euchromatin and heterochromatin, where euchromatin appears in a lighter shade and heterochromatin shades darker due to compact nucleosomal structures.
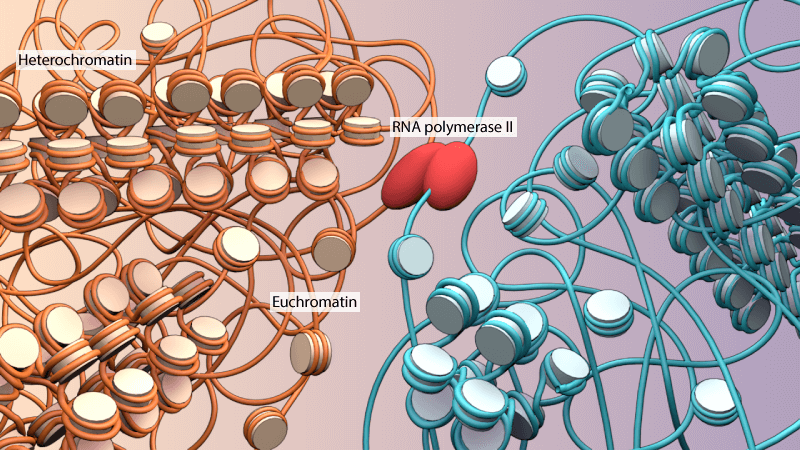
Euchromatin Functions
Euchromatin is loosely bound nucleosomes and exposed DNA helical sequences that carry active genes accessible for transcription by polymerases (DNA and RNA) and other regulatory protein complexes.
- The presence of euchromatin in a nucleus describes the function of transcriptionally active cells.
- And cells use the transformation mechanism from euchromatin to heterochromatin to regulate the level of gene expression and replications.
- Cells control the chromatin structure as needed; in the case of active genes or when genes are turned on, euchromatin structure can be observed.
- In ‘housing keeping genes’, euchromatin form is always present as they are constantly transcribed for the basic survival of cells.
- Many environmental interactions have led to post-translational modifications of histone octamer, causing alterations in the chromatin structure with fewer or no changes to the DNA sequence.
- The accelerated aging process can be caused due to epigenetic alterations showing increased euchromatin expression in the nucleus.
Different Genomic Regions of Euchromatin
Euchromatin possesses active genes containing many genetic sequences, such as gene bodies, promoters, and enhancers promoting transcription.
Transcriptionally Active Gene Bodies
- The gene bodies refer to the euchromatin transcript with introns and exons from start to end.
- Active histone modification is required to keep the gene bodies in an open chromatin state, allowing easy polymerase access.
- Gene bodies are enriched with DNA methylation in highly expressed genes to prevent intragenic transcription initiation.
- The nucleosomal position gets weaker along the length of active gene bodies promoting transcription.
Promoters
- As the name suggests, promoters are regions in a gene with a specific sequence or motifs read by polymerases and promote transcription initiation.
- They are present upstream of the 5’ end of the coding or sense strand of the transcribing gene, where the polymerase recognizes the sequence to initiate transcription.
- They are usually 100-1000 base pairs in length.
- Promoters show characteristic motifs of GC-rich DNA sequences. Vertebrate promoters have 70% GC nucleotide base pairs.
- Examples include TATA box, E-box (sequence CACGTG), etc.
Enhancers
- Enhancers are DNA sequences that regulate transcription by binding to the transcription factors.
- They can be present upstream or downstream from the respective gene and at various distances away from promoters.
- In an open state, the enhancer sequence may be far away from the coding strand, but in a folded state, it may be located near the transcription start site.
- The orientation of the enhancer sequence (forward or reverse) has no effect on the gene transcription.
Regulation of Euchromatin
There are many processes, such as methylation, acetylation, and phosphorylation, that regulate the formation of the euchromatin structure. The regulations are made on the histone protein by the recruitment of enzymes in post-translational modifications. Enzymes act on the N-terminal tail of nucleosomal histone, which can either form an open structure of euchromatin or a closed form of heterochromatin.
Acetylation and Methylation
- Histone acetylation leads to euchromatin formation, whereas histone methylation forms a heterochromatin structure.
- Acetylated histones are more negatively charged, disrupting the DNA helix with histones, making it more accessible for action by polymerases and transcription factors.
- Acetylation generally occurs on the lysine residues present in the N-terminal tails of histone protein, further increasing access to transcription factors.
Phosphorylation
- Euchromatin structures are also regulated by the addition and removal of phosphate groups by kinase and phosphatase enzymes.
- The phosphate group is added to the serine, tyrosine, or threonine residues on nucleosomal euchromatin.
- The addition of the phosphate group increased the negative charge of DNA, causing the helix to relax, hence opening the DNA strand for transcription.
Conclusion
Chromatin is categorized into euchromatin and heterochromatin based on nucleosomal positioning in the eukaryotic cells. Euchromatin structure is more transcriptionally active and less condensed than heterochromatin, which contains repressed DNA sequences. Euchromatin comprises repetitive units of DNA helix wrapped around histone proteins in a more open state to allow easy transcription. Euchromatin is specifically marked by histone modifications via methylation and acetylation to facilitate gene expression. Alterations in the chromatin structure due to environmental factors lead to the transformation of euchromatin to heterochromatin and vice-versa. Increased euchromatin presence is associated with an enhanced aging process suggesting higher transcription and expression of genes.
References
- Euchromatin – https://biologydictionary.net/euchromatin/
- Euchromatin – https://en.wikipedia.org/wiki/Euchromatin
- Morrison, Olivia, and Jitendra Thakur. “Molecular complexes at euchromatin, heterochromatin and centromeric chromatin.” International Journal of Molecular Sciences 22.13 (2021): 6922.
- Chemical Composition of Chromatin – https://www.biologydiscussion.com/chromosomes/eukaryotic-chromosomes/chemical-composition-of-chromatin/36090
- International Human Genome Sequencing Consortium. “Finishing the euchromatic sequence of the human genome.” Nature 431.7011 (2004): 931-945.
- Promoters – https://www.addgene.org/mol-bio-reference/promoters/
- Promoter (genetics) – https://en.wikipedia.org/wiki/Promoter_(genetics)
- Enhancer – https://www.nature.com/scitable/definition/enhancer-163/
