A gene is a sequence of DNA that codes for RNA or a Protein. It is the basic unit of hereditary that is passed from one generation to another. The gene determines the characteristics of the offspring.
Interesting Science Videos
What is a Gene?
- The term gene was coined by Danish botanist Wilhelm Johannsen.
- Gene is the basic functional unit of hereditary.
- The whole genome of an organism can be divided into genes.
- The genes code for proteins in a series of processes which are the building blocks of life.
- There are several types of genes with specific functions and positions.
- Within a genome, there are various coding and non-coding regions.
- The genomes of eukaryotes and prokaryotes are distinguished by the amount of non-coding regions present.
- The human genome is approximately 3,200 Mb large, and the genome of the most studied organism E. coli is 4.6 Mb large. (1 Mb = 1,000,000 bases).
- Mycoplasma genitalium has the smallest genome, which contains only 468 genes and is 0.58 Mb large.
- The genes can exist in different forms known as alleles.
Structure of a Gene
- The genes are located on the chromosomes at a specific location called the locus.
- The genes and the DNA are compactly packed in the chromosome.
- Each nucleated cell contains the whole set of the genome.
- In humans, the genome is composed of 23 pairs of linear chromosomes packed with the help of histone proteins.
- 22 pairs are of autosomal chromosomes and 1 pair consists of sex chromosomes.
- In bacteria like E. coli, the genome is composed of a single circular chromosome.
A typical Gene consists of:
- Promoter sequence: It is a sequence of DNA to which the enzyme binds which initiates the process of gene expression. It is present at the start of the gene.
- Coding region: It is a stretch of DNA that codes for proteins or RNA. The coding region is composed of introns and exons. Exons are the regions which are protein-coding sequences and intron sequences do not code for any protein. It is also called Cistron. Cistron consists of Muton (the part of a gene that undergoes mutation) and Recon (the part of a gene that undergoes recombination).
- Terminator sequence: It is the sequence of DNA that brings about the termination of the gene expression. It is present at the end of a gene.
Prokaryotic Gene
The prokaryotic gene consists of –
- A single promoter
- Coding region
- A single terminator
The gene in which a single promoter and terminator control the expression of many genes is known as the Poly-cistronic gene OR The gene which codes for one or more proteins is known as the polycistronic gene.
Promoter
- A promoter sequence is present in the upstream region of the gene (near 5`end of the gene). -35 (TTGACA) and -10 (TATAAT) are known promoter sequences that initiate the process of transcription by interaction with the RNA polymerase.
- -35 and -10 regions are consensus sequences, which means that they are conserved sequences in man organisms.
- The -10 region is also known as the Pribnow box.
Coding region
- This region starts with the initiator sequence and ends with the terminator sequence.
- The coding region is responsible for the formation of proteins by subsequent steps of transcription (formation of mRNA from DNA template) and translation (formation of the amino acid polypeptide chain and further into folded proteins from mRNA strand as a template).
- The prokaryotic gene is continuous, which means it does not contain introns (the non-coding region in a gene).
Termination region
- This region signals the RNA polymerase to terminate the transcription process.
- The termination can be of two types Rho (ρ) dependent or Rho (ρ) independent termination.
Examples
The genome of E.coli is well characterized and consists of 4,267 genes. The genome exists as circular, double-stranded DNA.
The genome of Mycoplasma genitalium contains only 468 genes.
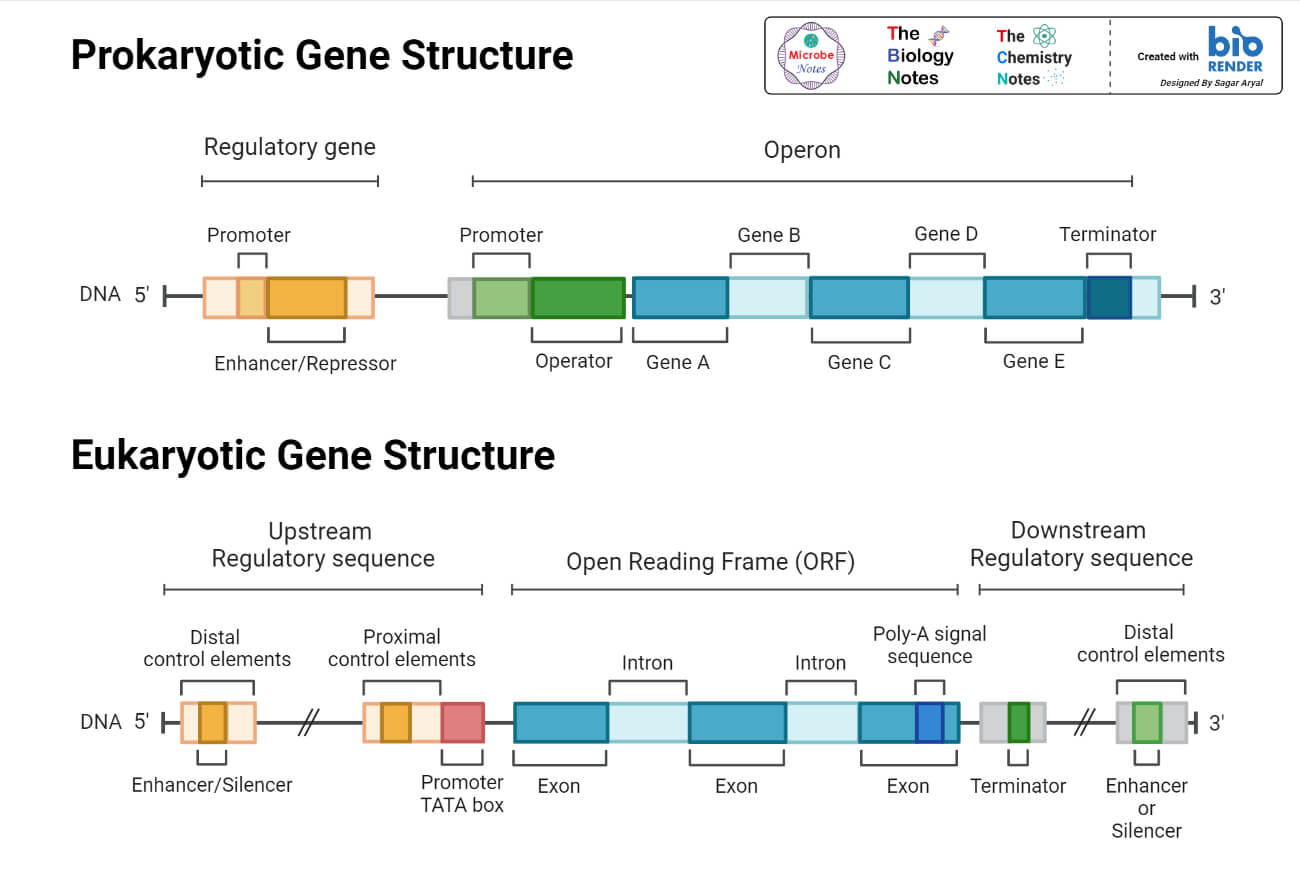
Eukaryotic Gene
The eukaryotic gene consists of –
- Exons
- Introns
- Promoter sequence
- Termination sequence
Exons
- The term exon was coined by Walter Gilbert in 1978.
- These are the coding sequences that are first transcribed and then translated which leads to the formation of proteins.
- The numbers of exons vary in an organism.
Introns
- Introns were discovered by Richard Roberts and Phil Sharp. Their experiments showed that the eukaryotic genes contain interruptions, called introns.
- These interruptions or introns do not code for any protein and hence, are also known as non-coding regions or junk segments of DNA.
- The introns are removed from the mRNA segment before it is translated into a protein by the process known as splicing.
- Introns are important as they are responsible for regulatory sequences of the RNA and regulate gene expression.
- Exon shuffling, in which introns facilitate the recombination of exons in different genes, is evolutionarily important.
Promoter sequences
- This is the region where the process of transcription is initiated.
- In eukaryotes, the promoter contains three distinct regions known as a core promoter, proximal promoter, and distal promoter.
- The core promoter is the site recognized by the RNA polymerase, and this region is located just before the start site. TATA box is the site that contains the sequence 5`-TATAA-3` and also has sites for histone binding and transcription factors.
- The proximal promoter site is located upstream of the core promoter and usually has binding sites for primary regulatory elements for the transcription process.
- A distal promoter is present upstream of the proximal promoter, and this promoter also has binding sites for transcription factors but mainly contains regulatory elements.
Termination sequence
- The RNA polymerase recognizes the particular sequence on the mRNA, which indicates the termination of the transcription process.
- In bacteria, the termination can be carried out in two ways. Ρ (rho) dependent and ρ (rho) independent termination.
- In ρ-dependent termination, the ρ enzyme is required. It binds to a Rut (Rho utilization site) site. This enzyme binds to a specific sequence which is C rich region. This binding cleaves the RNA from the template.
- In the ρ independent termination, a few nucleotides upstream of the termination site a G-C rich region is present and near the termination site also a G-C rich region is present. These two sites are complementary to each other. These sites form a hairpin loop structure and this formation of a hairpin loop drags the RNA from the template, and termination is achieved.
- In the case of the process of translation, the termination codons present on the mRNA indicate the termination. The termination codons are UAA, UGA, and UAG.
Regulation of gene expression
Gene expression is regulated for proper functioning and differentiation of the cells. Every cell contains a different set of proteins which are coded by the genes. Hence the regulation of gene expression is important for the organism.
Prokaryotic gene expression regulation
- In prokaryotes, the processes of transcription and translation occur almost simultaneously. When a particular protein is no longer required, the process of transcription stops, as the protein in excess amount signals the stop of the transcription process.
- Hence the expression of a gene in prokaryotes is mostly regulated at the transcriptional level.
- Three kinds of proteins regulate the genes. Inducers, Repressors, and Activators.
- Repressor proteins bind to the operator region and block the binding capacity of the RNA polymerase.
- In the case of activator proteins, they bind to the inducer region of the gene and enhance the binding capacity of the RNA polymerase.
- Inducer molecules bind to the DNA and activate or repress the gene based on the needs of the cell and the availability of substrate.
- In prokaryotes, the genes required for a particular protein are assembled next to each other, and this arrangement is called an operon.
- For example – The genes required for the use of lactose are arranged into lac operon. Trp (tryptophan) operon is a repressible operon.
- Bacteria require certain amino acids for survival, and these amino acids are produced in the cell with the help of these operons. Tryptophan is one of the essential amino acids required by bacteria. 5 genes are required for synthesizing tryptophan, and these genes are placed next to each other.
- If tryptophan is not available in the environment, bacteria synthesize tryptophan using these genes.
- When the concentration of tryptophan in the cell is high, two molecules of tryptophan bind to the repressor protein on the operator site and inhibit the transcription of RNA hence genes required for the synthesis of tryptophan are blocked.
- Trp operon is negatively regulated by tryptophan molecule.
- Another example of gene regulation in prokaryotes is the lac operon. This is an inducible operon.
- When the glucose concentration in a cell is less or glucose is absent, then bacteria can utilize lactose as a source of energy. Lac Z gene in the lac operon produces β-galactosidase, which breaks down lactose into galactose and glucose.
- When lactose is present, its isomer allolactose binds to the repressor protein and changes its structure so that it cannot bind to the lac operator site and prevent transcription.
- The presence of lactose induces the operon to code for proteins and enzymes and so it is an inducible operon.
- The absence of glucose and the presence of lactose are the important conditions required for a functional lac operon.
Eukaryotic gene expression regulation
- The gene expression in eukaryotes is much more complex as compared to prokaryotes. This is because the process of transcription takes place in the nucleus, and translation is carried out in the cytoplasm. Both processes are not carried out simultaneously.
- Eukaryotic gene expression can be regulated at various levels. The levels at which gene expression in eukaryotes is regulated are:
1. Chromatin remodeling/Epigenetic modification
- A particular region of the chromosome should be opened for the transcription process to take place and the binding of the transcription factors and enzymes.
- Histones pack the DNA into nucleosome complexes. Histone proteins can move along the DNA strand to expose a particular part of the DNA for transcription.
- When nucleosomes are closely placed, the gene cannot be transcribed, butt as the nucleosomes slide along the DNA, the transcription factors can bind to the DNA for initiation of transcription.
- Modification in histone proteins affects the nucleosome spacing.
- Various functional groups (methyl, phosphate, or acetyl groups) are attached to specific amino acids in the histone proteins (positively charged), which can affect nucleosome spacing in DNA.
- The addition of chemical modifications such as acetyl groups to histone proteins reduces the positive charge of the histone proteins resulting in weak binding of DNA with the histones. This creates certain open regions for transcription.
- DNA molecule is modified by DNA methylation, which occurs in specific regions known as CpG islands. This is CG rich region, and the cytosine in this region has the ability to bind with the methyl group.
- The modifications in chromatin organization interact with methylated regions of DNA.
- DNA methyltransferases are attracted to regions where histone proteins are modified.
- This highly methylated DNA with de-acetylated histones is coiled very tightly and hence inactive for transcription.
2. Transcriptional control
- This is the most common type of gene regulation in which the formation of mRNA is controlled.
- During transcription, various transcription factors bind to the promoter sequences. TATA box is one of the consensus sequence 5`- TATAAT-3` to which transcription factors containing TATA-binding proteins bind. This binding of transcription factors helps in binding the RNA polymerase to the promoter sequence for the transcription process.
- This controls the initiation of the transcription process.
- Other than promoter sequences, enhancer sequences are also present in a eukaryotic gene. These enhancer regions are usually present upstream of the gene and not close to the gene they enhance.
- Specific transcription factors bind to these enhancer regions.
- When a particular protein transcription factor binds to the enhancer site, the shape of the proteins changes so that it can interact with the proteins present at the promoter site.
- As the enhancer region is upstream of the promoter region, the structure of DNA has to be modified so that the enhancer region is close to the promoter region.
- To do so, the DNA bending proteins bend the DNA in such a manner that the proteins in the enhancer region can interact with the proteins in the promoter region.
- Every enhancer has short DNA sequences known as distal core elements.
- The activators at the distal core elements interact with the transcription factors.
- Some promoters may be observed in two different genes, but the distal core elements are different for every gene, and hence the gene expression is different and is regulated.
- There are transcriptional repressors as well which bind to the promoter or the enhancer region and repress the process of transcription.
3. Post-transcriptional control
- The post-transcriptional control involves the processing of pre-mRNA into mature mRNA.
- The pre-mRNA contains introns which are removed through the process of splicing, and the mature mRNA is obtained.
- One of the splicing mechanisms is alternative splicing, which involves the joining of exons in different patterns.
- The difference in the joining of exons leads to different proteins and is one of the gene expression regulation mechanisms.
- Once the mature mRNA is ready to be transported outside the nucleus, Poly G capping is done at the 5` end, and Poly A capping is done at the 3` end of the RNA. This prevents the mRNA from exonuclease activity and stabilizes the molecule.
- The life span of the RNA in the cytoplasm varies and can be controlled by different factors.
- Every RNA molecule has a different life span and it is degraded or it decays after a particular time.
- If the RNA resides in the cytoplasm for a longer time and has a lower decay rate, more protein will be formed. If the decay rate of the RNA molecule is increased, it will reside for a shorter time in the cytoplasm, and hence less protein will be formed.
- The decaying rate or decaying time of the RNA molecule depends upon the stability of the RNA.
- The RNA molecule contains un-translated regions or UTRs (these are different from introns). Certain proteins are known as RNA binding proteins (RBPS) bind to the UTR regions, and the binding of RBPs increases or decreases the stability of the RNA molecule in the cytoplasm.
- Other than RBPs, elements known as microRNAs also bind to the RNA molecule and have the ability to increase or decrease the stability of the RNA molecule.
4. Translational control
- The rate of initiation of the translation process is regulated by the binding of translational initiation factors.
- When eIF-2 is phosphorylated, there is a conformational change, and it cannot bind to GTP, and initiation of translation is blocked. If eIF-2 remains un-phosphorylated, then the initiation process can occur.
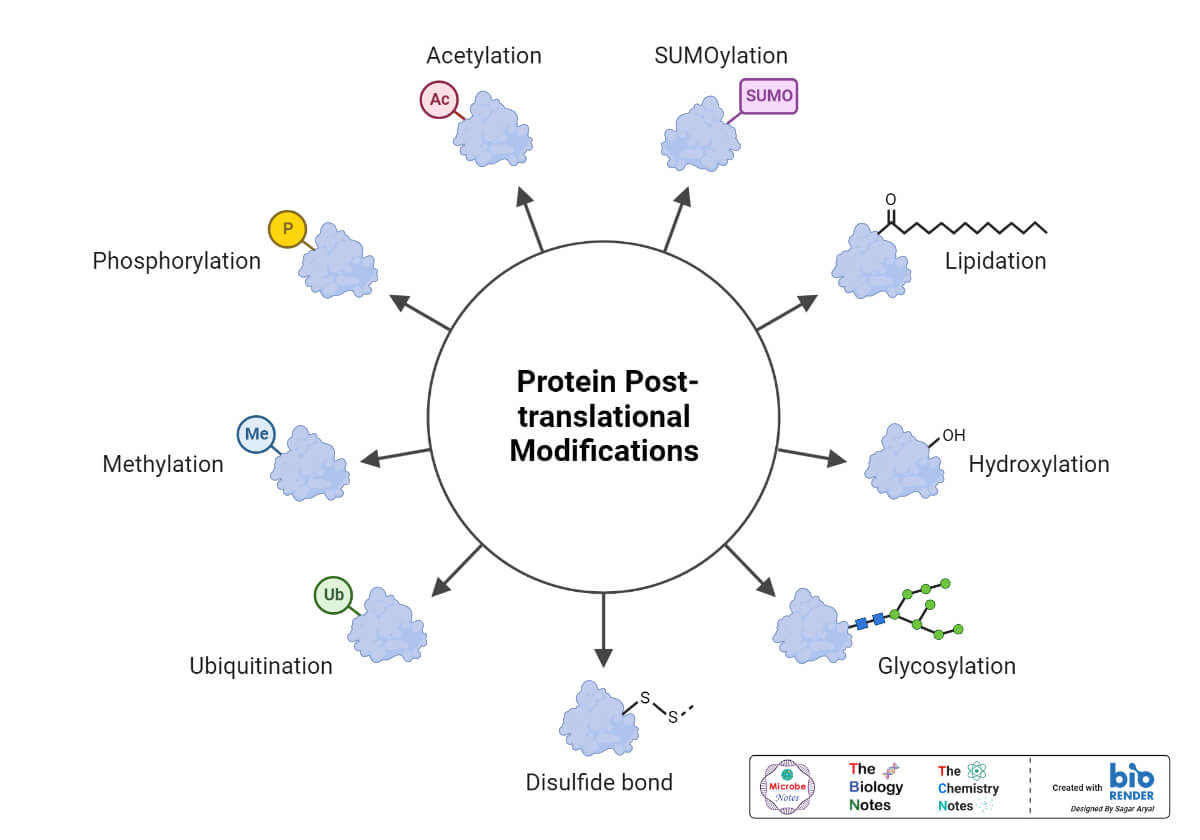
5. Post-translational modification
- This includes regulation of the modification of the immature protein into an active protein.
- The addition of chemical groups such as phosphate, acetyl, methyl, and ubiquitin affects the stability of proteins and also their activity.
- As proteins are involved in every step of gene expression and regulation, any modification in them affects other processes such as transcription, post-transcription events, translation, stability of the RNA, and also post-translational events.
- If a protein has ubiquitin attached to it, it undergoes degradation via a complex known as the proteasome.
Gene Mutations
An inheritable change in the nucleotide sequence of chromosomes is known as mutation.
OR
Mutation can be defined as a sudden and abrupt change occurring in the sequence of nucleotides of DNA in a cell.
Mutations can be broadly classified into Spontaneous mutations and Induced mutations.
Spontaneous mutations: These mutations occur under natural environmental conditions without the presence of any mutagenic agent.
Some reasons for spontaneous mutations are
- Errors in DNA replication
- Exposure to various mutagenic agents such as UV rays, cosmic rays, radioactive compounds, etc.
- Tautomerism of natural nitrogenous bases, A and C change from stable amino to imino, and G and T change from stable keto to enol.
Induced mutations: These mutations result when an organism is exposed to a known mutagenic agent such as radiation or various chemicals that react with DNA.
The process of inducing mutations is known as mutagenesis.
Different classes of mutations are as follows –
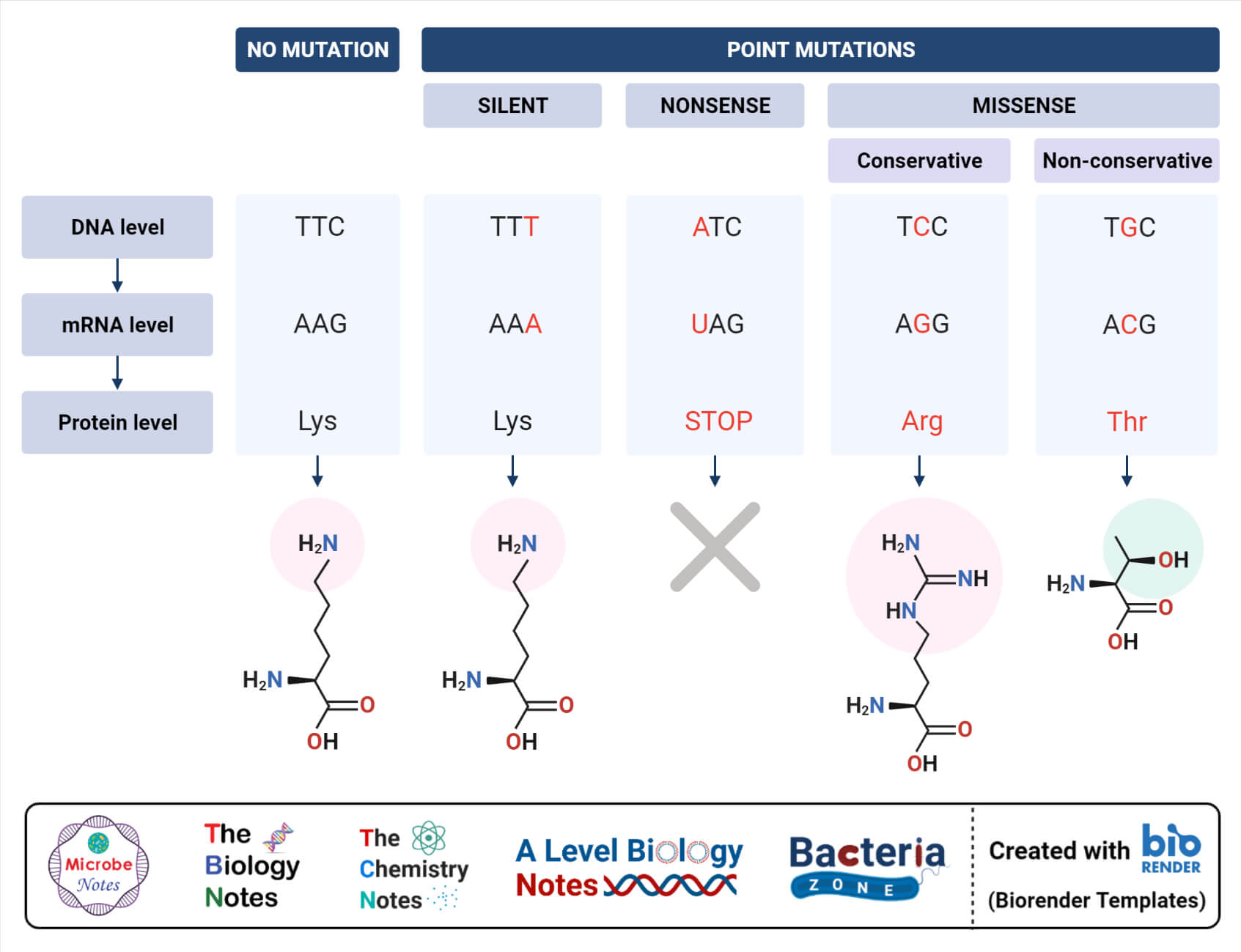
A) Point mutations
1. Base pair substitution – In this type of mutation, one base pair is incorrectly added during the process of replication and replaces the pair on the corresponding position on the complementary strand.
Based on which pair is substituted, there are two subtypes as
Transversion – In this, a purine base is replaced by a Pyrimidine base and vice versa.
Transition – In this, a Purine base is substituted by a different Purine base, and Pyrimidine is replaced by another Pyrimidine base.
Missense mutation – In this type of mutation, the altered codon codes for non-functional protein.
Non-sense mutation – In this type of mutation, the codon is altered to termination or non-sense codon hence protein synthesis is terminated.
Example – sickle cell anemia is caused due to substitution mutation.
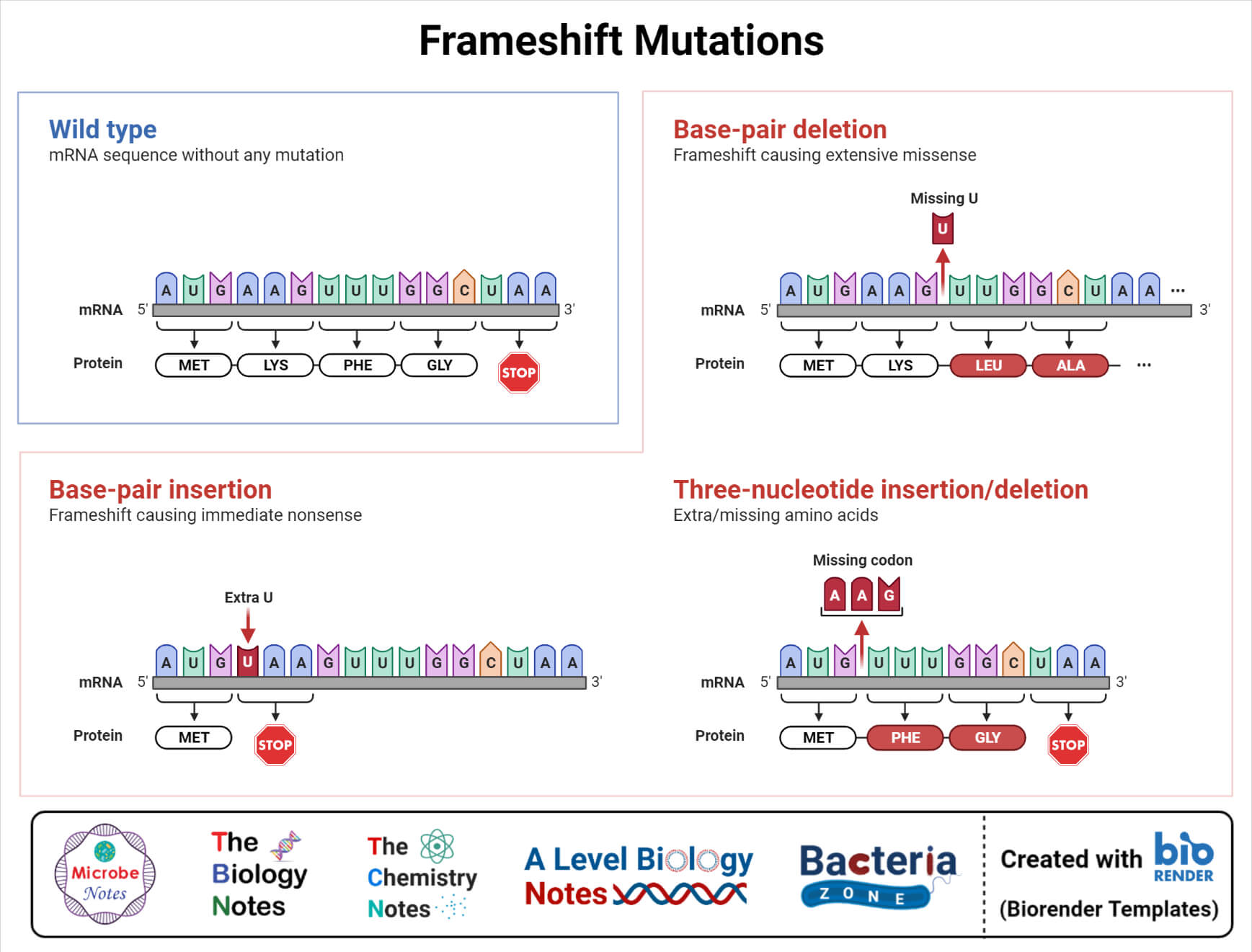
2. Insertion – In this type of mutation, one or more than one nucleotide are added to the replicating strand, which often results in a frameshift of the open reading frame.
Example – one form of beta-thalassemia is caused due to insertion mutation.
3. Deletion – In this type of mutation, one or more than one nucleotides are excised or skipped during replication which often results in a frameshift.
Example – cystic fibrosis is caused due to deletion mutation.
B) Chromosomal mutations
1. Inversion – In this type of mutation, the part of the chromosome is inverted and then reinserted.
Example – Hemophilia A is caused due to inversion of the chromosomal region.
2. Deletion – in this type of mutation, a region of the chromosome is lost, which results in the absence of all of the genes in that particular region.
Example – Duchenne muscular dystrophy is caused due to deletion of a region in the chromosome.
3. Duplication – In this type of mutation, a region of the chromosome is repeated, which results in the extra production of proteins from the repeated genes.
Example – some cancers are caused due to duplication of genes.
4. Translocation – in this type of mutation, a region of the chromosome is abnormally attached to another chromosome.
Example – one form of leukemia is caused due to translocation.
C) Copy number variation
1. Gene amplification – in this type of mutation, the number of tandem copies of the locus is increased.
Example – breast cancers are caused due to gene amplification mutation.
2. Expanding tri-nucleotide repeats – In this type of mutation, the normal number of repeated tri-nucleotides sequences is expanded.
Example – Huntington’s disease, fragile X chromosome is caused due to expanding of tri-nucleotides sequences.
Gene Inheritance
Gene inheritance is the process by which genetic information is passed on from one generation or parent generation to the next generation or off-springs.
Gene inheritance in humans
- In humans, X and Y are the sex chromosomes that are responsible for the inheritance of genetic information to the next generation.
- Females have XX chromosomes, and males have XY chromosomes.
- The inherited gene is responsible for the genotype and phenotype of the offspring.
- Genotype is the genetic makeup of the offspring and is unique for every individual. The genotype describes the entire genome and genes of the individual.
- Phenotype is the physical description of the individual. Phenotype depends on the genotype hence the physical characteristics are also unique for every individual.
- During mating, the egg released by females has an X chromosome; the perm of males either carries an X or Y chromosome.
- If the X chromosome is inherited by sperm, the offspring is a female as the mating of egg and sperm results in XX chromosomes.
- If the Y chromosome is inherited by sperm, the offspring is a male as the mating of egg and sperm results in XY chromosomes.
- There are two major types of gene inheritance – Single gene inheritance and Multiple gene inheritance.
- In single gene inheritance, the same chromosome is inherited from the father and mother. The genes may be the same or alleles of each other.
Example – genes for eye color, height, etc
- In multiple gene inheritance, multiple genes or alleles are inherited for the same genotype and phenotype.
Example – ABO blood group system alleles in which IA IA and IO IA both represent blood group A.
Type of inheritance patterns
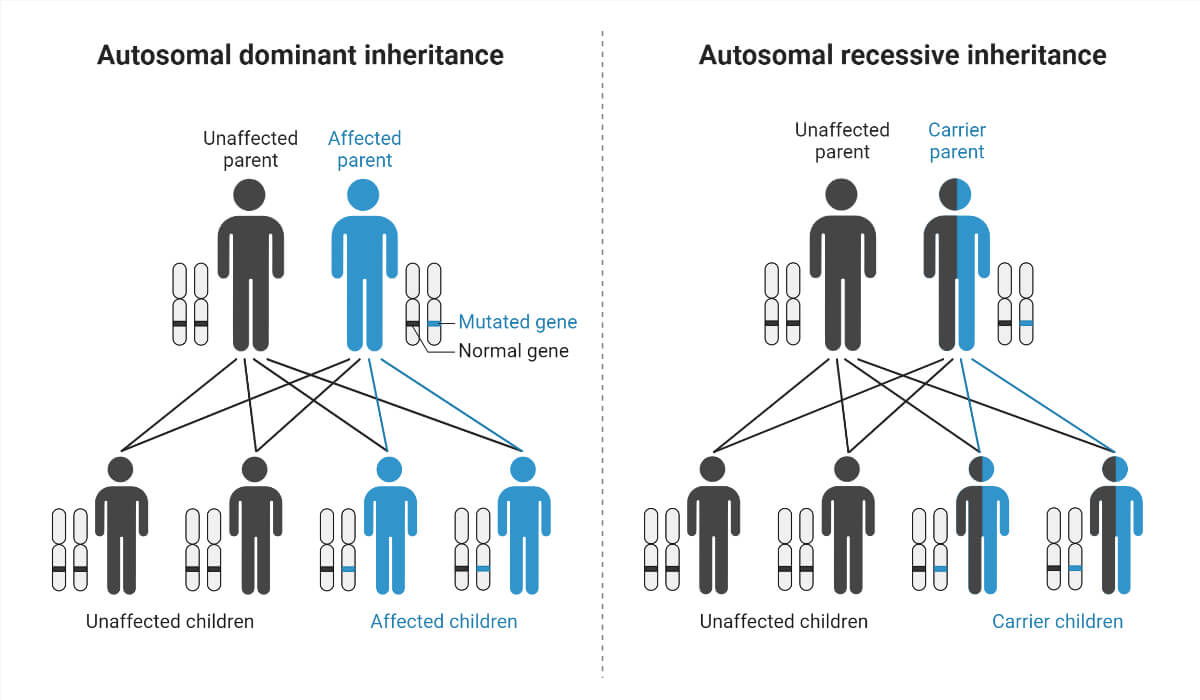
- Autosomal dominant inheritance – Autosomal inheritance is referred to the traits inherited by autosomes. Humans have 22 pairs of autosomes. Dominant gene in the autosomes is inherited, leading to an Autosomal dominant condition.
- Autosomal recessive inheritance – Autosomal recessive is the inheritance of recessive genes by autosomes.
- X-linked recessive inheritance – Humans have a pair of sex chromosomes. In females, the sex chromosomes are XX, and in males, XY. The X-linked inheritance is the one in which genes are inherited and are present on the X chromosome. When a recessive gene is inherited, the inheritance is X-linked recessive inheritance.
- X-linked dominant inheritance – When a dominant gene is inherited via the X chromosome, it is X-linked dominant inheritance.
- Mitochondrial inheritance – This includes the inheritance of genes in the mitochondria. Mitochondria are present only in egg cells and not in sperm cells. Hence mitochondrial inheritance occurs only via egg cells.
- Multifactorial inheritance – This includes the inheritance of genes from both the parents as well as other factors, such as environmental factors that influence the inheritance.
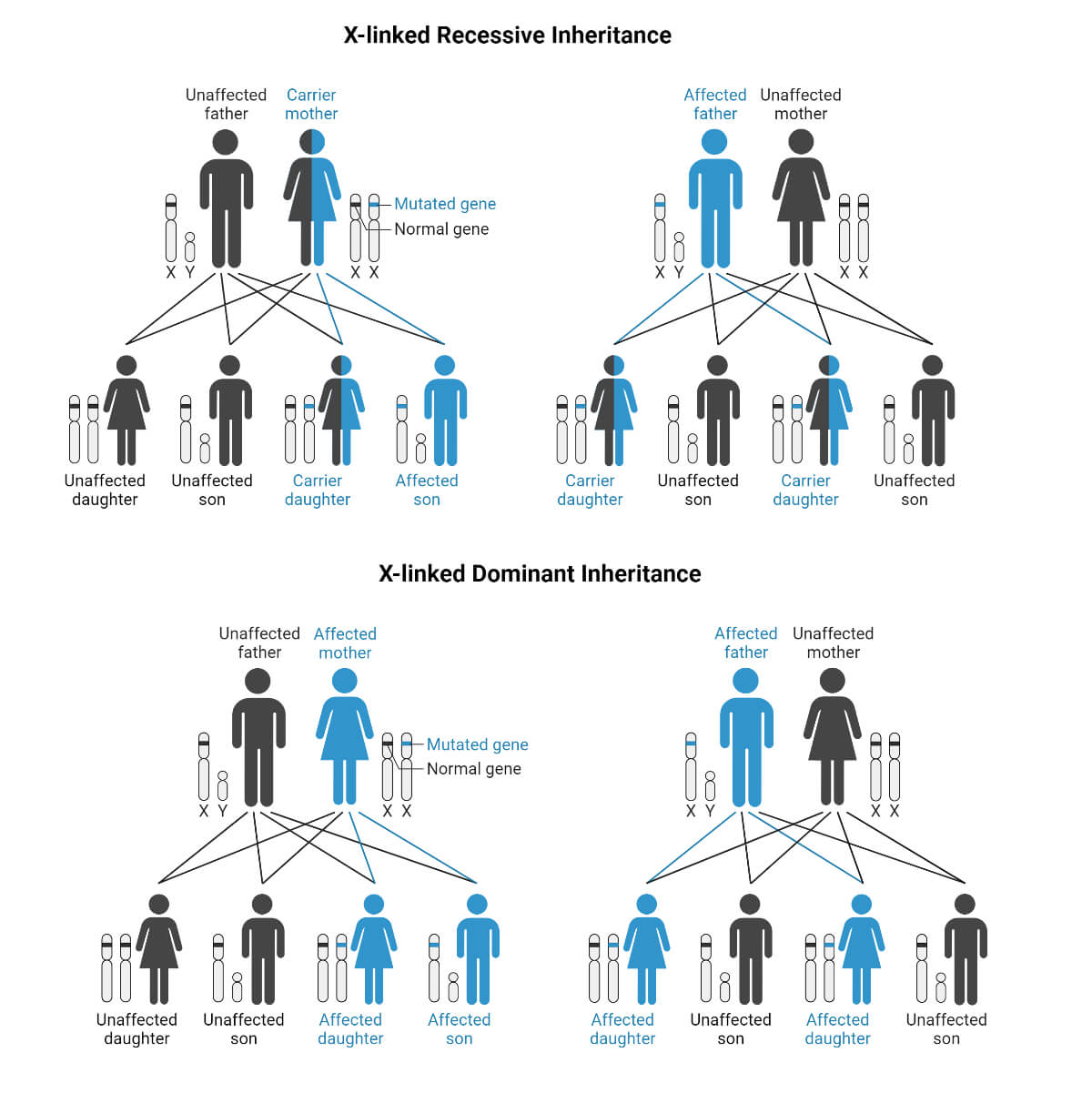
Gene inheritance in prokaryotes
Vertical gene transfer
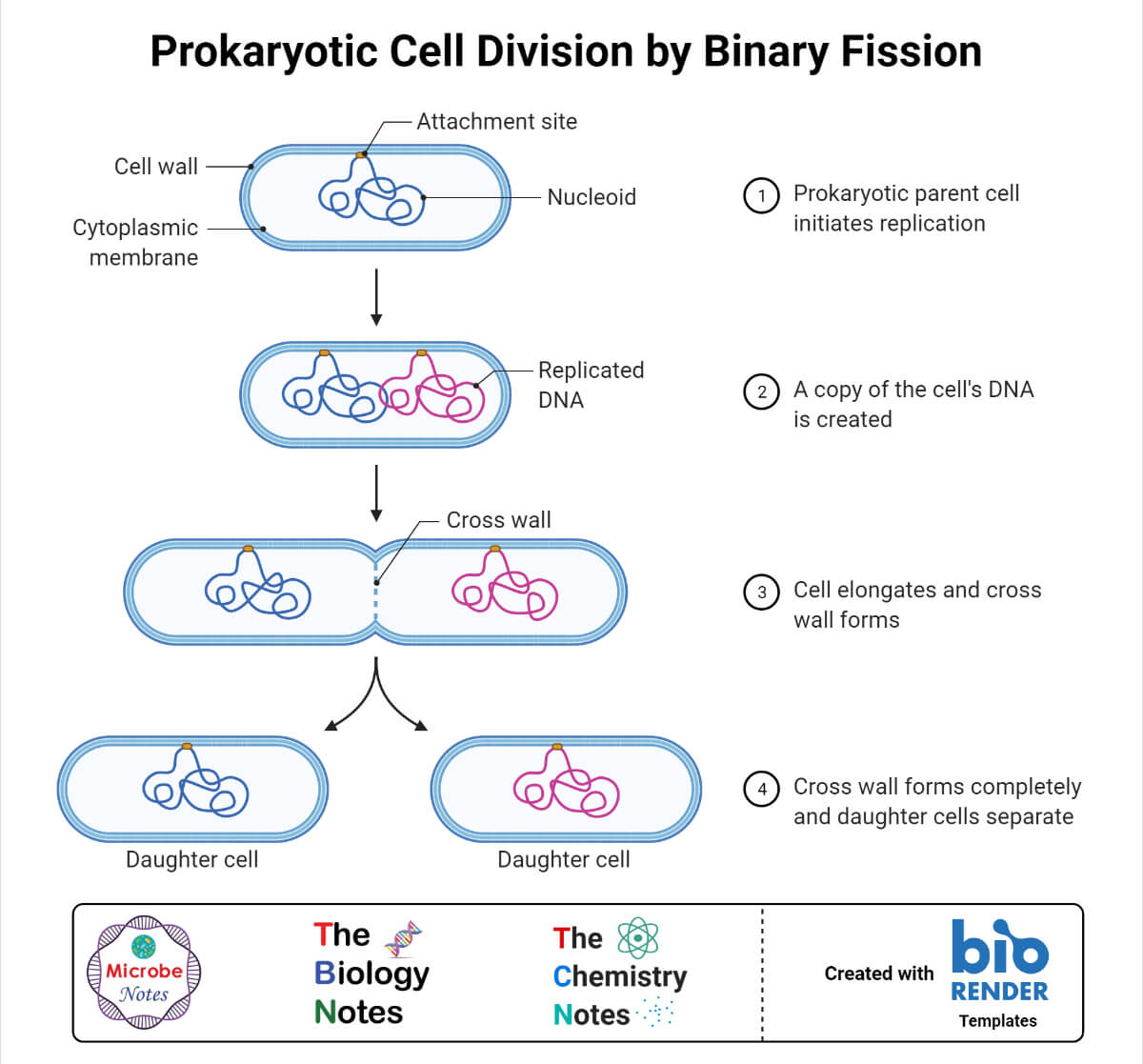
- In prokaryotes, genes are transferred from one generation to another when the cell divides through binary fission. This is known as vertical gene transfer.
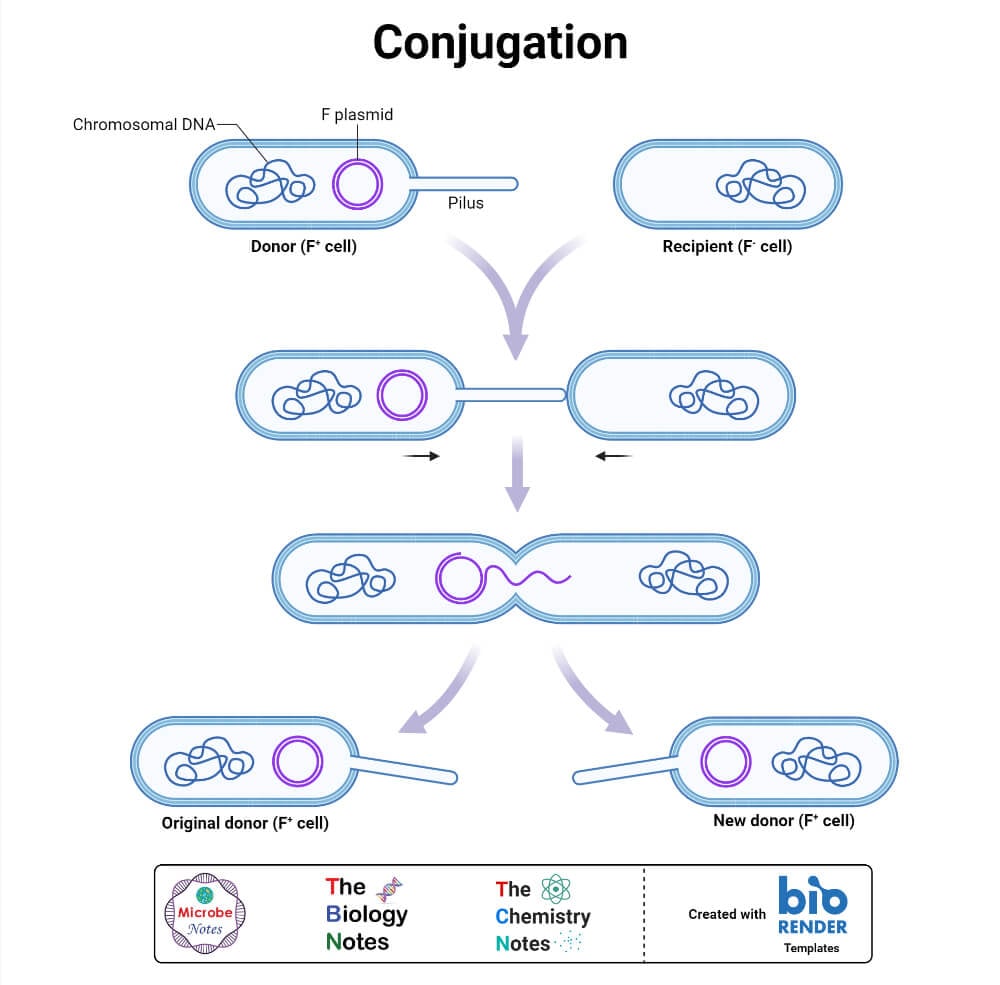
- Horizontal gene transfer is the process by which some genes are transferred from one cell to another (which is not an offspring). Or gene transfer amongst adjacent cells.
- The three mechanisms of horizontal gene transfer are – transformation, conjugation, and transduction.
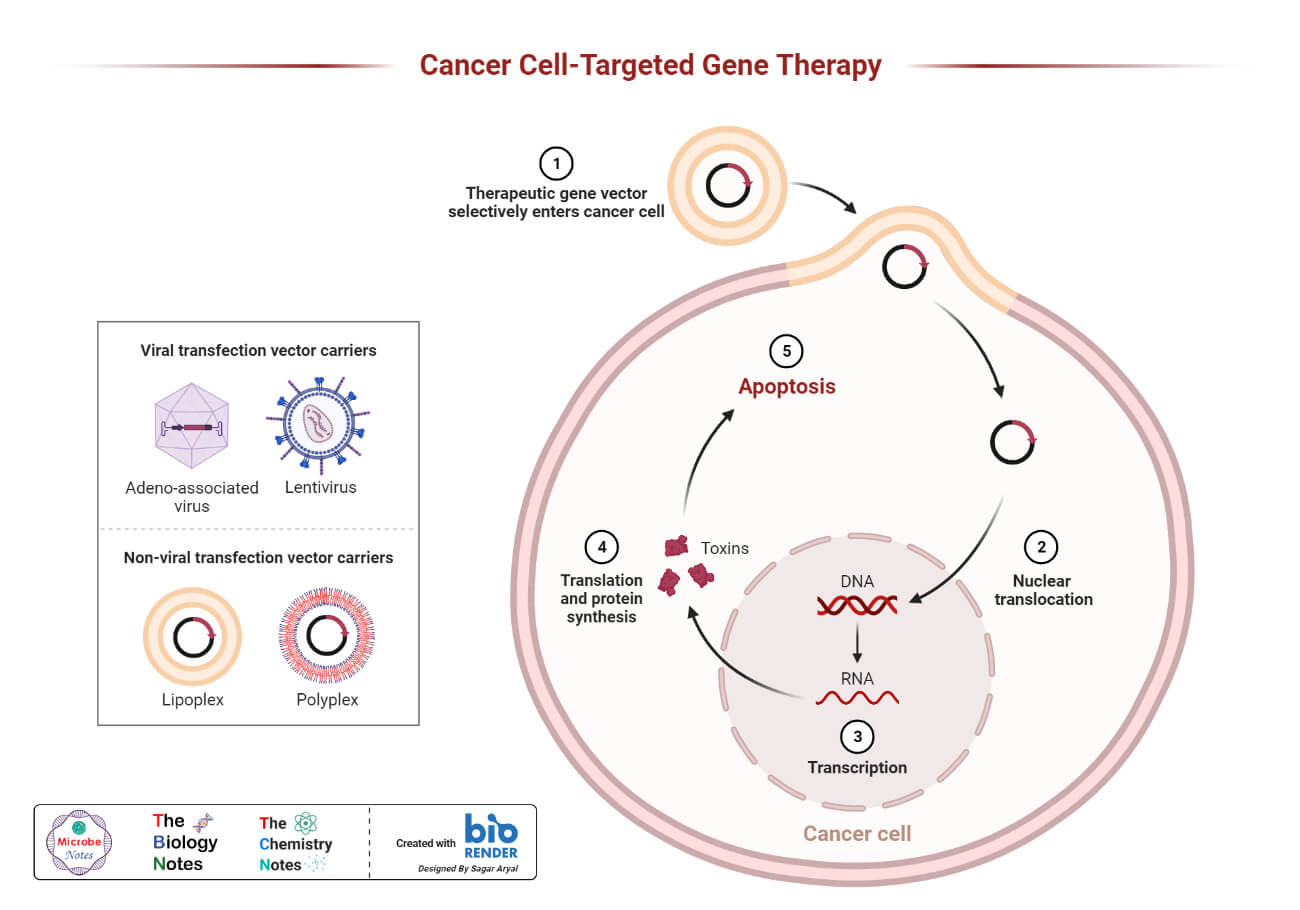
Gene Therapy
Gene therapy is the process by which genes are altered to treat or stop a particular disease.
Gene therapy can be divided into types.
1. Somatic gene therapy – This approach of gene therapy involves the alteration or correction of mutated genes of the somatic target cells, and the altered gene will not be inherited to the next generation.
2. Germline gene therapy – This approach involves the alteration or correction of mutated genes that are inherited to the next generation.
- Gene therapy widely uses recombinant DNA technology in which the gene of interest or a healthy gene is inserted in a specific vector, which later releases the gene in the target cell of the organism.
- The vectors used for gene transfer are usually plasmids or viruses, which are modified so that the virus does not have any virulence capacity or does not infect the organism.
- Other vectors such as stem cells or liposomes are also used for the transfer of a particular gene.
- Examples of vectors – retrovirus, adenovirus, herpes simplex virus, plasmids etc.
Advantages of gene therapy
- Various mutations or deletions in genes can be replaced using gene therapy.
- The genetic disease which cannot be cured using medicines can be cured, and this prevents the inheritance of the disease to the next generation.
Disadvantages of gene therapy
- There is a possibility of unwanted immune responses while using gene therapy.
- The vector used may regain its virulence ability and can cause harmful diseases.
- Along with the target cell, the new gene inserted in the individual may target other cells as well.
- Gene therapy is relatively costly as compared to other treatment options.
References
- Brown TA. Genomes. 2nd edition. Oxford: Wiley-Liss; 2002. Chapter 1, The Human Genome. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21134/
- Polyak K, Meyerson M. Overview: Gene Structure. In: Kufe DW, Pollock RE, Weichselbaum RR, et al., editors. Holland-Frei Cancer Medicine. 6th edition. Hamilton (ON): BC Decker; 2003. Available from: https://www.ncbi.nlm.nih.gov/books/NBK12983/
- https://www.genome.gov/25520306/online-education-kit-1977-introns-discovered
- https://www.addgene.org/mol-bio-reference/promoters/
- https://opentextbc.ca/biology2eopenstax/chapter/prokaryotic-gene-regulation/
- https://www.nature.com/scitable/topicpage/genetic-mutation-441/
- https://www.nature.com/scitable/topic/gene-inheritance-and-transmission-23/
- https://mydoctor.kaiserpermanente.org/ncal/specialty/genetics/resources/inheritance.jsp
- Wirth, T., Parker, N., & Ylä-Herttuala, S. (2013). History of gene therapy. Gene, 525(2), 162–169. https://doi.org/10.1016/j.gene.2013.03.137
- Hall Rebecca J., Whelan Fiona J., McInerney James O., Ou Yaqing, Domingo-Sananes Maria Rosa: Horizontal Gene Transfer as a Source of Conflict and Cooperation in Prokaryotes . Frontiers in Microbiology Volume 11, Year 2022.
- https://www.frontiersin.org/article/10.3389/fmicb.2020.01569
