DNA molecules most commonly exist in double-stranded form whereas RNA exists in single-stranded form. However, both DNA and RNA when in single-stranded form may exist to form a hairpin loop-like structure called stem-loop or hairpin structure. Hairpin loop (secondary structural folds) is observed both in Ribonucleic acid (RNA) and Deoxyribonucleic acid (DNA) but mostly, it is observed in RNA.
Interesting Science Videos
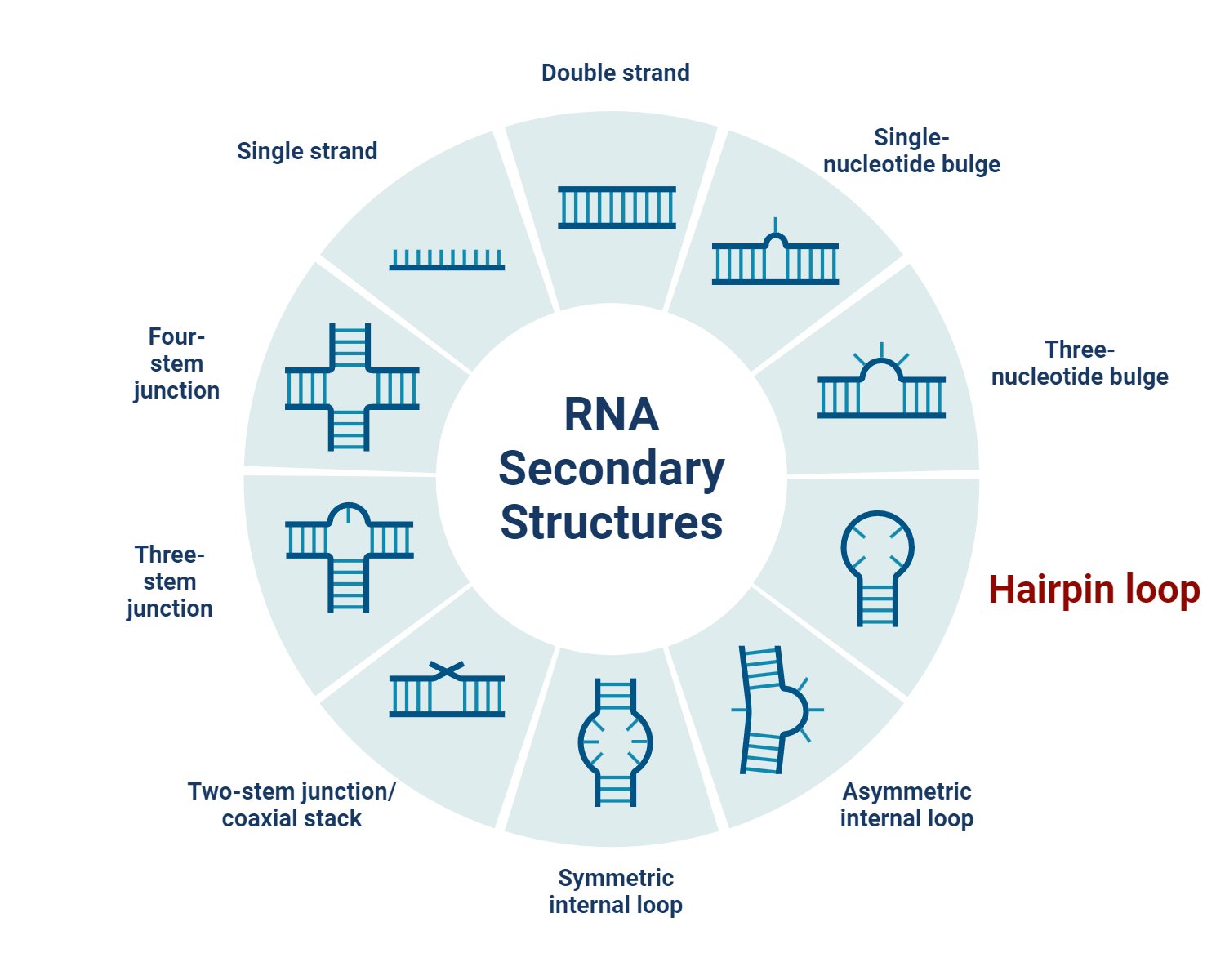
What is a stem-loop (hairpin loop)?
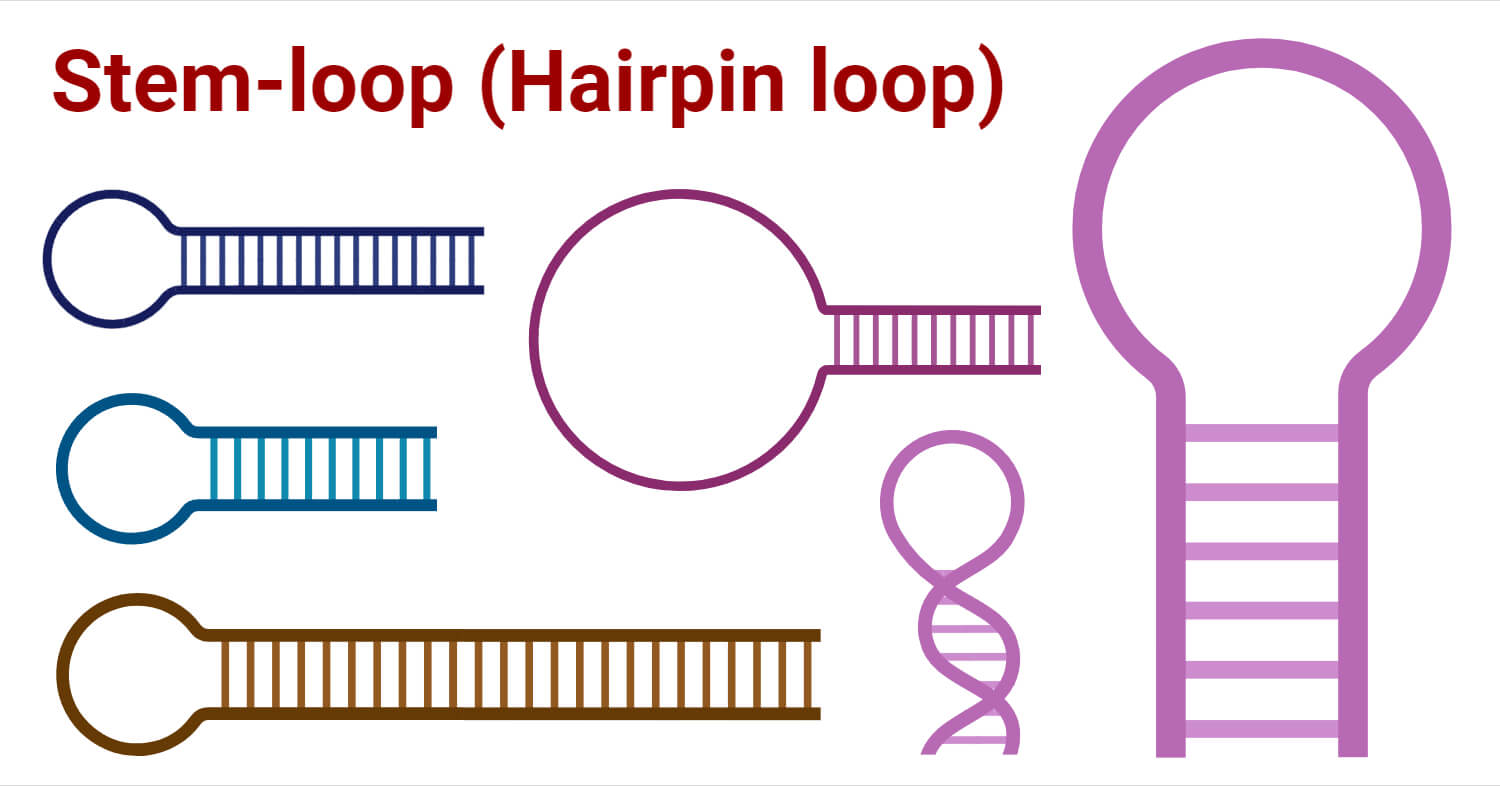
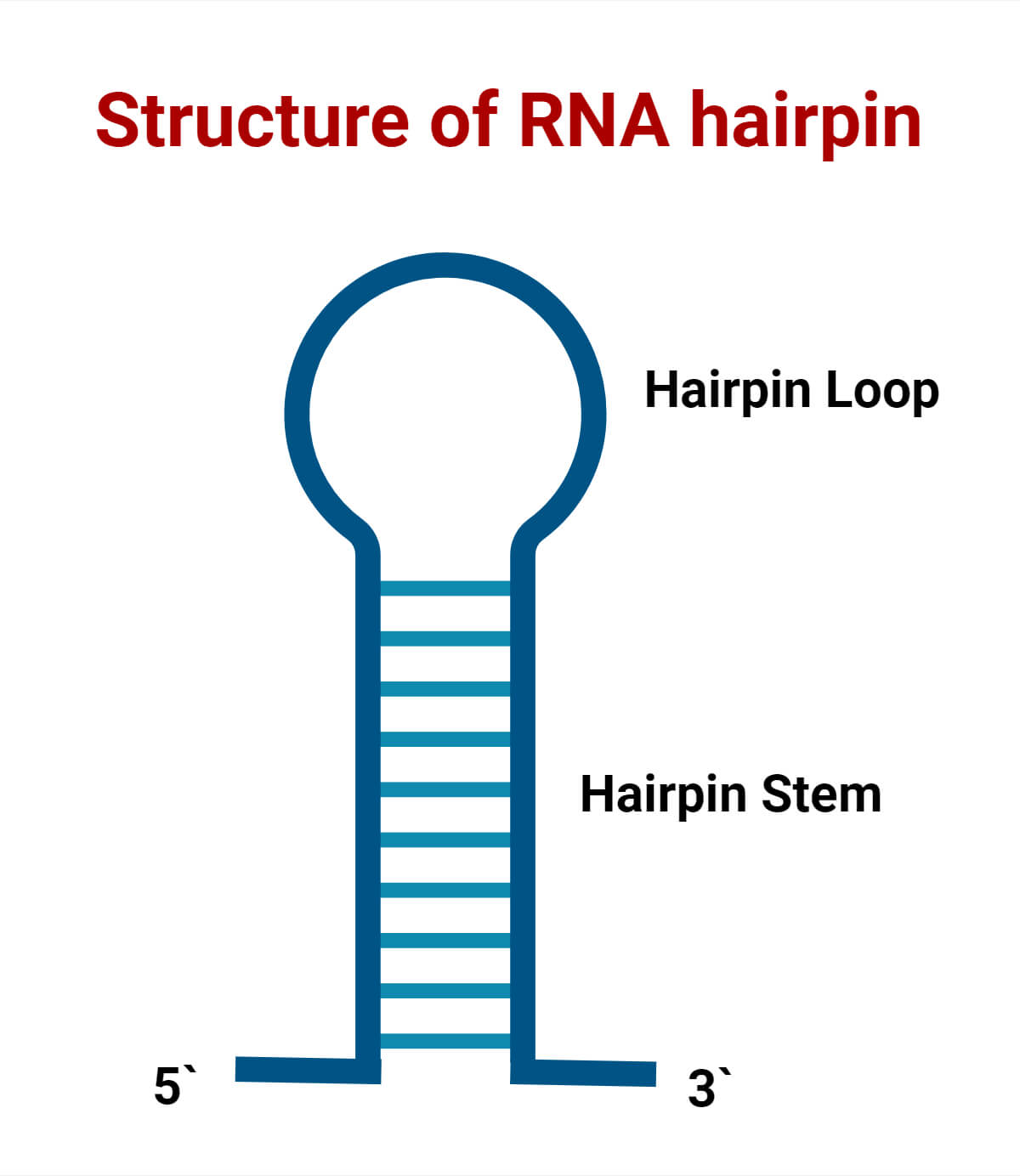
A Stem-loop can be defined as a hairpin-like pattern formed because of the intramolecular base pairing of the nucleotide sequence (especially in palindromic sequence) within the same molecule. Here, the nucleotide pairs within the molecule to form a double helix structure ending in an unpaired loop resulting in a hairpin-like structure.
Characteristics of stem-loop (hairpin loop)
- Hairpin loops form in single-stranded nucleic acids and consist of a base-paired stem and a loop sequence with unpaired nucleotide bases.
- They consist of several unpaired bases within the loop, varying from 7 to 20 base pairs(bp).
- It is often described by the number of unpaired bases within the loop, with prefixes like tri-, tetra-, penta-, etc., combined with the term “loop.”
- Hairpin loops form complementary regions within the same strand base-pair, creating a double helix ending in an unpaired loop. Their structure in nucleic acid consists of a base-paired stem structure and a loop sequence with unpaired or non-Watson-Crick-paired nucleotides. The resulting structure looks like a loop or a U-shape.
- In RNA, mRNA molecules can form hairpin loops when two complementary sequences bind together after folding.
- Hairpin loops with a central GNA trinucleotide motif (G, guanine; N, any nucleotide; A, adenine) have been found to form particularly stable structures. For example, for the sequence 5′-GCGCAGC, a melting transition for disruption of the hairpin structure of 67°C has been reported.
Note: Hairpin loop formation also occurs in DNA, rapidly, typically within microseconds, beyond current maximum molecular dynamics simulation timescales.
Origin of RNA hairpin
An RNA hairpin comprises a double-stranded RNA (dsRNA) stem with mismatches and bulges, along with a terminal loop, making it a prevalent secondary structure in RNA folding predictions.
RNA hairpins originate via two mechanisms:
- Transcription by DNA-dependent RNA polymerase of inverted repeat DNA sequences, leading to RNA folding into hairpin loop structures.
- Formation of RNA molecule as a template for RNA-dependent RNA polymerase, synthesizing the second stem strand. This mechanism, observed mainly in certain viral replication processes, produces long, perfect dsRNA hairpins, often in a ‘copy-back’ replication mechanism.
Note: RNA hairpins regulate gene expression in cis or trans:
In cis: They regulate the same RNA molecule where they’re located.
In trans: They influence other RNAs or pathways beyond their origin.
Molecules with loop structures:
- Stem loops are integral components of RNA structures like pre-microRNA, transfer RNA (tRNA), RNA pseudoknots, and ribozymes.
- In tRNA, the anticodon recognizing a codon resides on an unpaired loop, while RNA pseudoknots feature nested stem-loop structures.
- Ribozymes such as the hammerhead ribozyme rely on stem-loop structures, with three stem-loops facilitating the self-cleavage activity.
- Hairpin loops within prokaryotic 5’UTRs often modulate translation or interact with regulatory proteins.
Stability Factors of stem-loop (hairpin loop)
- The stability of stem-loop structures hinges on the stability of their helix and loop regions, controlled by various factors.
- Base pair stability is influenced by sequence, length, mismatches, and base composition, with guanine-cytosine pairings being more stable than adenine-uracil pairings.
- RNA commonly features guanine-uracil pairings, while base stacking interactions aid in helix stability.
- Loop stability is critical, with loops shorter than three bases being impossible and large, structure-less loops being unstable.
- Optimal loop length typically ranges from 4 to 8 bases, with sequences like UUCG forming particularly stable tetraloops, facilitating rapid formation.
Examples of stem-loop (hairpin loop)
- Hairpin loops are prevalent phenomena in nucleic acid strands, occurring in various instances.
- In prokaryotes, one example of a hairpin loop is the termination sequence for transcription, where encountering this loop causes the polymerase to disengage, hence terminating the transcription.
- Another notable example is tRNA, a key component in protein synthesis, which incorporates hairpin loops into its structure.
- Specifically, the tRNA molecule features three hairpin loops, resembling a three-leafed clover. Among these loops, one contains a crucial sequence known as the anticodon, responsible for recognizing and decoding mRNA molecules during translation.
- This clover-leaf configuration facilitates the precise alignment between codons, anticodons, and corresponding amino acids, crucial for accurate protein synthesis.
The palindromic DNA sequence —CCTGCXXXXXXXGCAGG— can form the following hairpin structure
—C G—
C G
T A
G C
C G
X X
X X
X X
X
A longer example of a 5‘ → 3‘ sequence of RNA that would lead to a hairpin loop structure is:
GCCGCGGGCCGAAAAAACCCCCCCGGCCCGCGGC
(Example source: https://www.bionity.com/en/encyclopedia/Stem-loop.html)
Significances of stem-loop (hairpin loop)
Significance of RNA Stem-loop Structure:
- RNA stem-loop structures are valuable for real-time measurements and biosensor fabrication, as properly designed stem-loop structured oligonucleotide probes can open in the presence of complementary sequences, enabling detection of unlabeled biomolecule targets like nucleic acids and proteins.
- In prokaryotic rho-independent transcription termination, mRNA hairpin loop formation prompts the dissociation of RNA polymerase from the DNA template.
- They act as binding sites for diverse proteins.
- They function as substrates for enzymatic reactions.
- They exhibit intrinsic enzymatic activities.
- Short hairpins within mRNA participate in diverse processes such as mRNA localization, translation regulation, and stability maintenance.
- Stem-loops contribute to translation initiation through internal ribosomal entry sites and play roles in viral replication.
- In RNA molecules, hairpin loops act as nucleation sites for RNA folding, impacting RNA-protein recognition and gene regulation.
- Stem-loop structures, like molecular beacons, enable specific detection of target sequences, providing opportunities for innovative diagnostic sensors and therapeutic methods.
DNA Hairpin Significances
- Single-stranded phages extensively utilize DNA hairpins throughout their life cycles, including at the origin of replication of E. coli.
- Cruciform DNA, featuring hairpin motifs, has been observed at various genomic locations, such as the RCR dso and N4 phage promoters.
- In eukaryotes, proteins that bind to cruciform DNA have been discovered, suggesting their involvement in genome translocation and replication initiation.
- Molecular Biology and Biotechnology: Hairpin DNA oligomer probes are essential tools in biotechnology and diagnostics due to their specificity in probe/target hybridization, aiding in match-versus-mismatch discrimination.
References
- Farahani, N., Behmanesh, M., & Ranjbar, B. (2020). evaluation of Rationally Designed Label-free Stem-loop DnA probe Opening in the Presence of miR-21 by circular Dichroism and fluorescence techniques. Scientific reports, 10(1), 4018. https://www.nature.com/articles/s41598-020-60157-5
- Hairpin loop. Biology Online. Retrieved from https://www.biologyonline.com/dictionary/hairpin-loop
- Hairpin Loop. Creation wiki. Retrieved from https://creationwiki.org/Hairpin_loop
- Hairpin Loop. Eterna Wiki. Retrieved from https://wiki.eternagame.org/wiki/Hairpin_Loop
- Kannan, S., & Zacharias, M. (2007). Folding of a DNA hairpin loop structure in explicit solvent using replica-exchange molecular dynamics simulations. Biophysical journal, 93(9), 3218-3228 .https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2025651/
- RNA stem-loop. NIST. Retrieved from https://www.nist.gov/image/rna-stem-loop-0
- Stem-loop. BIONITY.COM. Retrieved from https://www.bionity.com/en/encyclopedia/Stem-loop.html
- Svoboda, P., & Cara, A. D. (2006). Hairpin RNA: a secondary structure of primary importance. Cellular and Molecular Life Sciences CMLS, 63, 901-908.https://www.nature.com/scitable/definition/hairpin-loop-mrna-314/#:~:text=A%20hairpin%20loop%20is%20an,secondary%20structure%20in%20RNA%20molecules.
- Swadling, J. B., Ishii, K., Tahara, T., & Kitao, A. (2018). Origins of biological function in DNA and RNA hairpin loop motifs from replica exchange molecular dynamics simulation. Physical Chemistry Chemical Physics, 20(5), 2990-3001.https://pubs.rsc.org/en/content/articlepdf/2018/cp/c7cp06355e
- Zhang, Y. (2013). Hairpin Structure. In: Dubitzky, W., Wolkenhauer, O., Cho, KH., Yokota, H. (eds) Encyclopedia of Systems Biology. Springer, New York, NY. https://doi.org/10.1007/978-1-4419-9863-7_325https://link.springer.com/referenceworkentry/10.1007/978-1-4419-9863-7_325
