- The Davson–Danielli model (or paucimolecular model) was a model of the plasma membrane of a cell, proposed in 1935 by Hugh Davson and James Danielli.
- The model describes a phospholipid bilayer that lies between two layers of globular proteins and it is trilaminar and lipoproteinous.
- It was the first model that attempted to describe the position of proteins within the lipid bilayer found in membranes.
- Danielli and Davson proposed a model whereby two layers of protein flanked a central phospholipid bilayer.
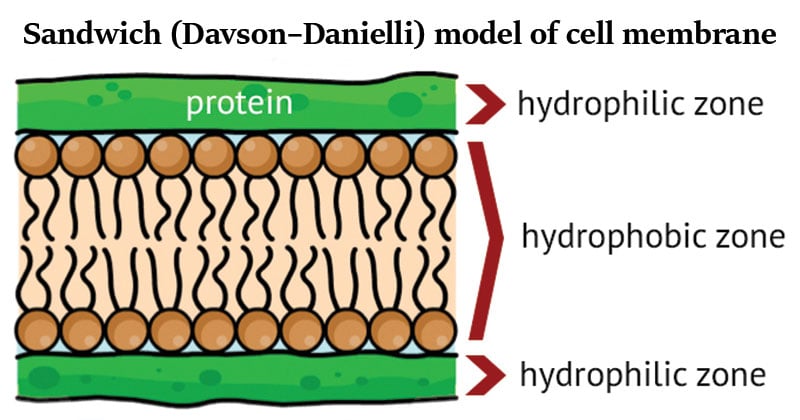
- The model was also described as a ‘lipo-protein sandwich’, as the lipid layer was sandwiched between two protein layers.
- The Davson–Danielli model predominated until Singer and Nicolson advanced the fluid mosaic model in 1972.
- The fluid mosaic model expanded on the Davson–Danielli model by including transmembrane proteins and eliminated the previously-proposed flanking protein layers that were not well-supported by experimental evidence.
Interesting Science Videos
Key Features of the Davson–Danielli Model
- Danielli and Davson, proposed a model, called sandwich model, for membrane structure in which a lipid bilayer was coated on its either side with hydrated proteins (globular proteins).
- Hence, the plasma membrane might be composed of two lipid-protein bilayers—one facing the interior of the cell and the other facing the external milieu.
- In this arrangement, the association between the surface proteins and bimolecular lipid leaflet would be maintained primarily by electrostatic interactions between the polar ends of each lipid molecule and charged amino acid side chains of the polypeptide layers.
- Either electrostatic or van der Waals bonds could bind other groups to the outer protein surface.
- Danielli and Davson proposed that such a membrane would exhibit selective permeability, being capable of distinguishing between molecules of different size and solubility properties and also between ions of different charge.
- From the speed at which various molecules penetrate the membrane, they predicted the lipid bilayer to be about 6.0 nm in thickness, and each of the protein layers of about 1.0 nm thickness, giving a total thickness of about 8.0 nm.
- A protein-lipid sandwich
- Lipid bilayer composed of phospholipids (hydrophobic tails inside, hydrophilic heads outside)
- Proteins coat the outer surface
- Proteins do not permeate the lipid bilayer.
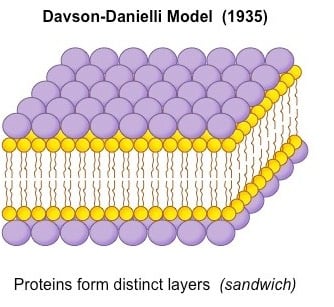
Image Source: BioNinja.
Support of Davson–Danielli model
- The Danielli-Davson model got support from electron microscopy.
- In high magnification electron micrographs, membranes appeared as two dark parallel lines with a lighter colored region in between.
- Proteins appear dark in electron micrographs and phospholipids appear light – possibly indicating proteins layers either side of a phospholipid core.
- The total thickness of the membranes too turned out to be about 7.5 nm.
Problems in the Davson–Danielli Model
- It assumed all membranes were of a uniform thickness and would have a constant lipid-protein ratio.
- It assumed all membranes would have symmetrical internal and external surfaces (i.e. not bifacial)
- It did not account for the permeability of certain substances (did not recognize the need for hydrophilic pores)
- The temperatures at which membranes solidified did not correlate with those expected under the proposed model.
Falsification Evidence for the Davson–Danielli Model
- Membrane proteins were discovered to be insoluble in water (indicating hydrophobic surfaces) and varied in size. Such proteins would not be able to form a uniform and continuous layer around the outer surface of a membrane.
- Fluorescent antibody tagging of membrane proteins showed they were mobile and not fixed in place. Membrane proteins from two different cells were tagged with red and green fluorescent markers respectively. When the two cells were fused, the markers became mixed throughout the membrane of the fused cell. This demonstrated that the membrane proteins could move and did not form a static layer (as per Davson-Danielli).
- Freeze fracturing was used to split open the membrane and revealed irregular rough surfaces within the membrane.
- These rough surfaces were interpreted as being transmembrane proteins, demonstrating that proteins were not solely localized to the outside of the membrane structure.
In light of these limitations, a new model was proposed by Seymour Singer and Garth Nicolson in 1972. According to this model, proteins were embedded within the lipid bilayer rather than existing as separate layers. This model, known as the fluid-mosaic model, remains the model preferred by scientists today (with refinements).
References
- http://ib.bioninja.com.au/standard-level/topic-1-cell-biology/13-membrane-structure/membrane-models.html
- https://biology.stackexchange.com/questions/20264/why-was-the-davson-danielli-model-rejected
- Lodish, H. F., Berk, A., Kaiser, C., Krieger, M., Scott, M. P., Bretscher, A., Ploegh, H. L., Matsudaira, P. T. (2008). Molecular cell biology. New York: W.H. Freeman.
- Smith, C. M., Marks, A. D., Lieberman, M. A., Marks, D. B., & Marks, D. B. (2005). Marks’ basic medical biochemistry: A clinical approach. Philadelphia: Lippincott Williams & Wilkins.
- https://www.revolvy.com/page/Davson%E2%80%93Danielli-model
- https://sebiology.weebly.com/blog/plasma-membrane-the-davson-danielli-model-vs-the-singer-nicolson-model
- http://www.oxfordreference.com/view/10.1093/oi/authority.20110810104725251
- http://yourdescribeinfo.blogspot.com/2018/05/davson-danielli-model.html
- http://www.library.armstrong.edu/eres/docs/eres/BIOL11071_NIXON/Membranes%20&%20Transport%20[Compatibility%20Mode].pdf
- http://psystems.disco.unimib.it/download/BiancoPhDThesis.pdf
- http://www.esalq.usp.br/lepse/imgs/conteudo_thumb/Cell-membrane-structure.pdf
- http://www.wou.edu/~guralnl/gural/102Chapter%2004%20-%20Plasma%20Membrane.pdf
- https://nptel.ac.in/courses/102103012/module1/lec3/2.html
- http://www.biologydiscussion.com/cell-biology/the-danielli-davson-membrane-model-with-diagram/3789
- https://www.researchgate.net/figure/Evolution-of-the-membrane-models-during-the-XX-th-Century-A-the-Davson-Danielli-model_fig1_317307719

Thank you very much
Thank you very much. It was well elaborated.
thank you for this I will b using this in my theory of knowledge essay <3
best