Real-Time PCR is a technique used to monitor the progress of a PCR reaction in real-time.
At the same time, a relatively small amount of PCR product (DNA, cDNA or RNA) can be quantified.
Real-Time PCR is based on the detection of the fluorescence produced by a reporter molecule which increases, as the reaction proceeds.
- Real-Time PCR is also known as a quantitative polymerase chain reaction (qPCR), which is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR).
- qPCR is a powerful technique that allows exponential amplification of DNA sequences.
- A PCR reaction needs a pair of primers that are complementary to the sequence of interest. Primers are extended by the DNA polymerase.
- The copies produced after the extension, so-called amplicons, are re-amplified with the same primers leading thus to exponential amplification of the DNA molecules.
- After amplification, however, gel electrophoresis is used to analyze the amplified PCR products and this makes conventional PCR time consuming; since the reaction must finish before proceeding with the post-PCR analysis. Real-Time PCR overcomes this problem.
- The term “real-time” denotes that it can monitor the progress of the amplification when the process is going on in contrast to the conventional PCR method where analysis is possible only after the process is completed.
PCR Terminology
| Polymerase chain reaction | PCR |
| Reverse transcription-polymerase chain reaction | RT-PCR |
| Real-time polymerase chain reaction | qPCR |
| RT-PCR / qPCR combined technique | qRT-PCR |
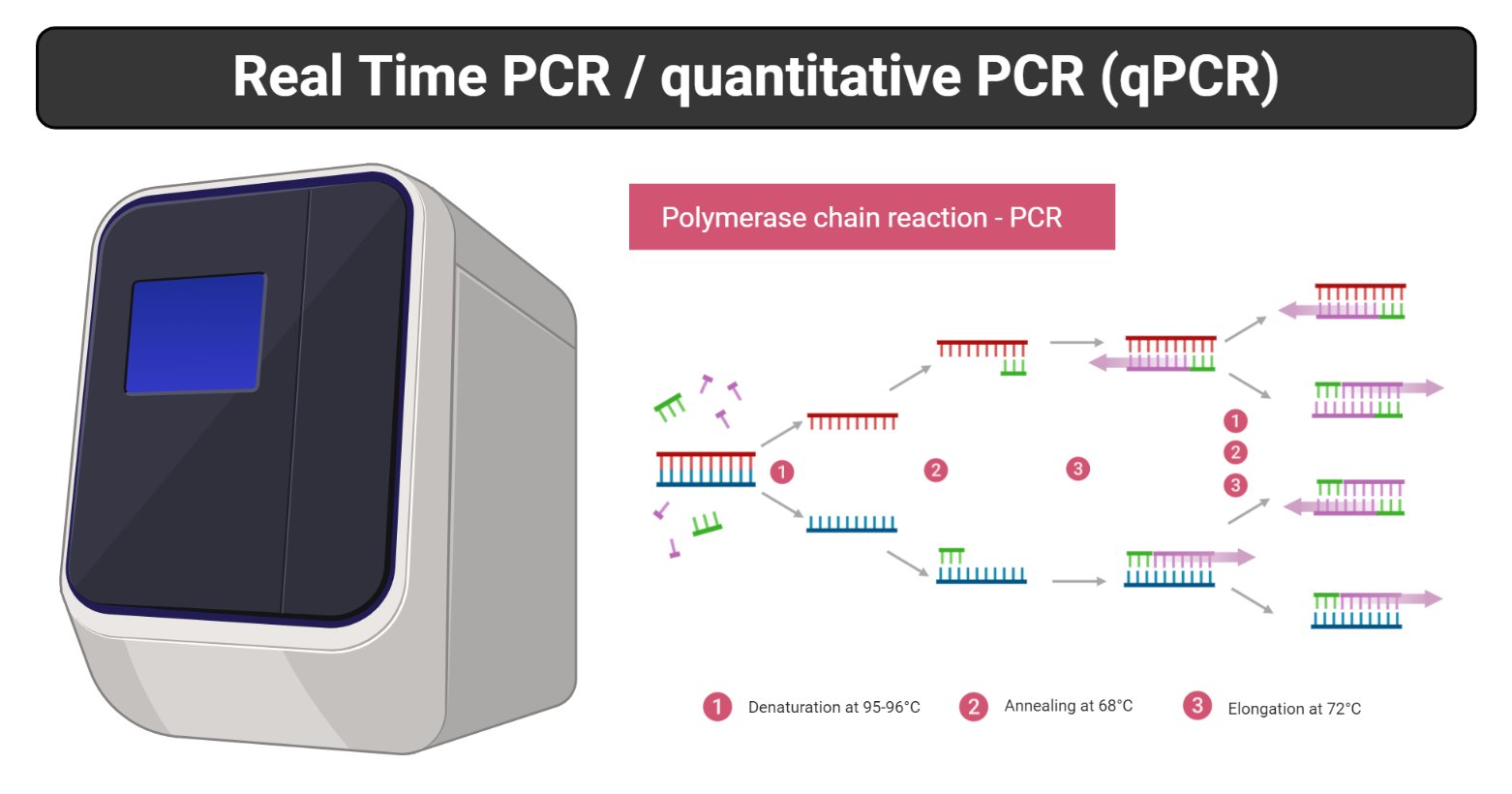
Image created with biorender.com
Interesting Science Videos
Principle of Real-Time PCR
This same principle of amplification of PCR is employed in real-time PCR. But instead of looking at bands on a gel at the end of the reaction, the process is monitored in “real-time”. The reaction is placed into a real-time PCR machine that watches the reaction occur with a camera or detector.
Although many different techniques are used to monitor the progress of a PCR reaction, all have one thing in common. They all link the amplification of DNA to the generation of fluorescence which can simply be detected with a camera during each PCR cycle. Hence, as the number of gene copies increases during the reaction, so does the fluorescence, indicating the progress of the reaction.
Steps of Real-Time PCR (Protocol)
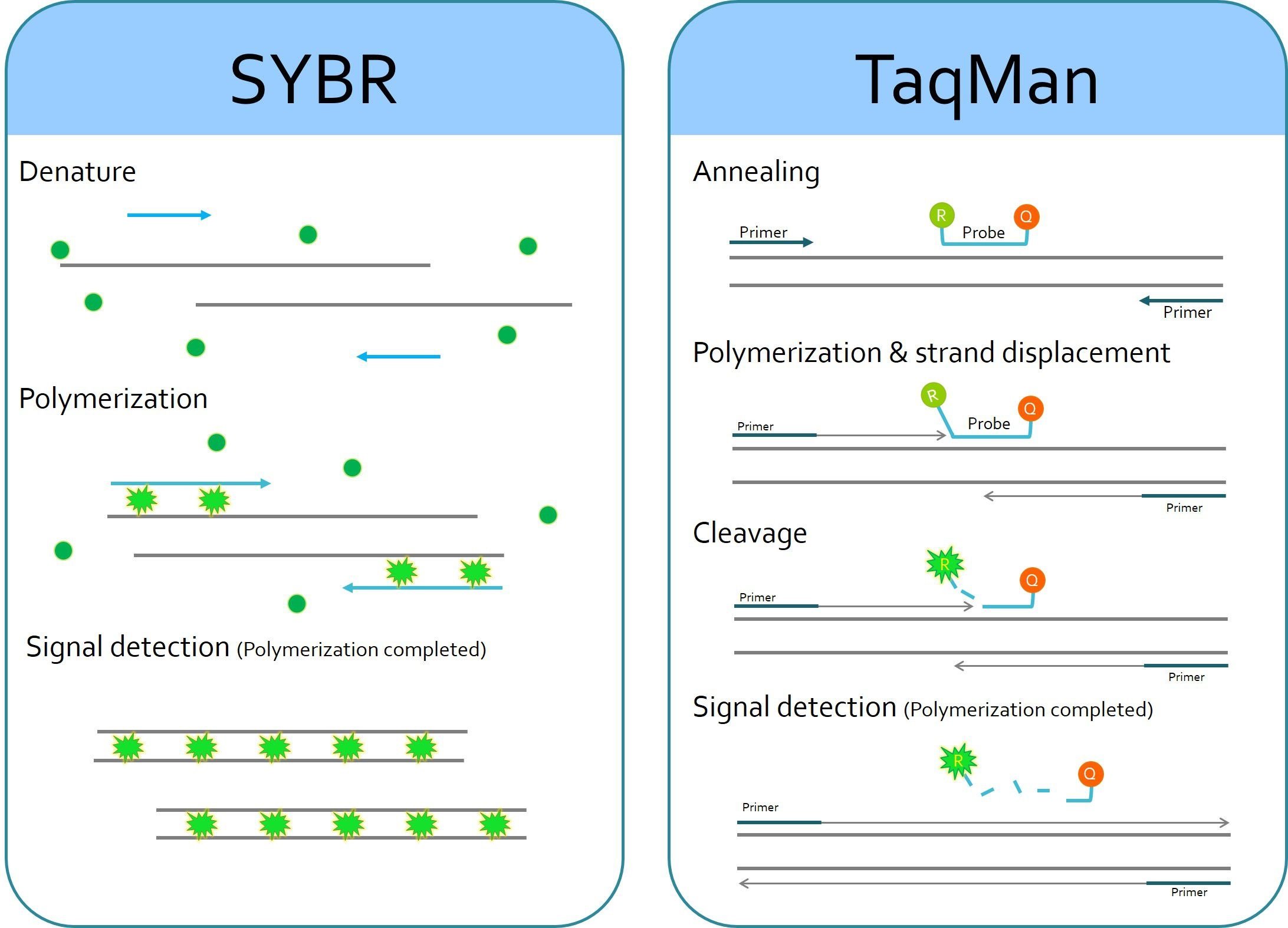
Image Source: SMOBIO Technology, Inc.
The working procedure can be divided into two steps:
A. Amplification
- Denaturation
High temperature incubation is used to “melt” double- stranded DNA into single strands and loosen secondary structure in single-stranded DNA. The highest temperature that the DNA polymerase can withstand is typically used (usually 95°C). The denaturation time can be increased if template GC content is high.
- Annealing
During annealing, complementary sequences have an opportunity to hybridize, so an appropriate temperature is used that is based on the calculated melting temperature (Tm) of the primers(5°C below the Tm of the primer).
- Extension
At 70-72°C, the activity of the DNA polymerase is optimal, and primer extension occurs at rates of up to 100 bases per second. When an amplicon in real-time PCR is small, this step is often combined with the annealing step using 60°C as the temperature.
B. Detection
- The detection is based on fluorescence technology.
- The specimen is first kept in proper well and subjected to thermal cycle like in the normal PCR.
- The machine, however, in the Real Time PCR is subjected to tungsten or halogen source that lead to fluoresce the marker added to the sample and the signal is amplified with the amplification of copy number of sample DNA.
- The emitted signal is detected by an detector and sent to computer after conversion into digital signal that is displayed on screen.
- The signal can be detected when it comes up the threshold level (lowest detection level of the detector).
Fluorescence Markers used in Real Time PCR
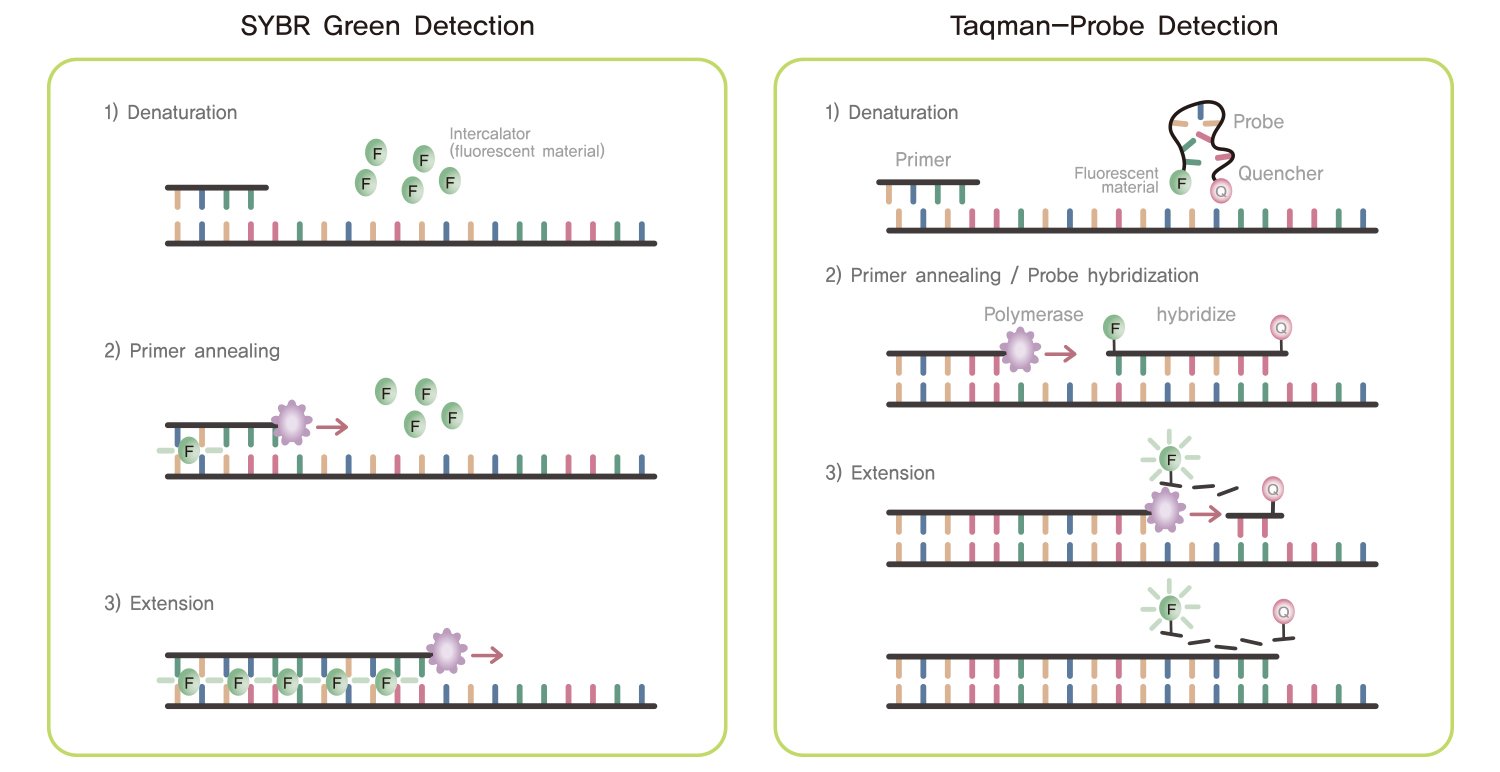
There are many different markers used in Real Time PCR but the most common of them include:
- Taqman probe.
- SYBR Green.
Taqman Probe
- It is a hydrolysis probe which bear a reporter dye, often fluorescein (FAM) at its 5’ end and a quencher tetramethylrhodamine (TAMRA), attached to the 3’ end of the oligonucleotide.
- Unders normal conditions, the probe remain coiled on itself bringing the fluorescence dye near the quencher, which inhibits or quenches of fluorescent signal of the dye.
- The oligonucleotide of the Taqpolymerase has a homologous region with the target gene and thus when the target sequence is present in the mixture, it bind with the sample DNA.
- As the taqpolymerase start to sunthesize new DNA strand in the extension stage, it causes degradation of the probe by 5’ end nuclease activity and the fluorescein is separated from the quencher as a result of which fluorescence signal is generated.
- As this procedure continues, in each cycle the number of signal molecule increases, causing the increase in fluorescence which is positively related with the amplification of the target.
SYBR Green
- This is a dye that emits prominent fluorescent signal when it binds at the minor groove of DNA, nonspecifically.
- Other fluorescent dyes like Ethidium Bromide or Acridine Orange can also be used but SYBR Green is better used for its higher signal intensity.
- SYBR Green is more preferred than the Taqman Probe as it can provide information about each cycle of amplification as well as about the melting temperature which is not obtained from the Taqman probe.
- However, its disadvantage is the lack of specificity as compared to Taqman Probe.
Advantages of Real-Time PCR
It has many advantages over the normal PCR:
- It gives a look in to the reaction that is help to decide which reactions have worked well and which have failed.
- The efficiency of the reaction can be precisely calculated.
- There is no need to run the PCR product out on a gel after the reaction as the melt curve analysis serve the purpose.
- The real-time PCR data can be used to perform truly quantitative analysis of gene expression. In comparison, old fashioned PCR was only ever semi-quantitative at best.
- Faster than normal PCR.
- Less complexity at the quantification of sample.etc.
Thus, unlike the ordinary preparative PCR, Real Time PCR allows the success of multiple PCR reaction to be determined automatically after only a few cycles, without separate analysis of each reaction, and avoids the problem of “false negatives”.
Applications of Real-Time PCR
- Gene expression analysis
- Cancer research
- Drug research
- Disease diagnosis and management
- Viral quantification
- Food testing
- GMO food
- Animal and plant breeding
- Gene copy number
References
- https://www.mun.ca/biology/scarr/Principle_of_RT-PCR.html
- http://www.primerdesign.co.uk/assets/files/beginners_guide_to_real_time_pcr.pdf
- https://link.springer.com/chapter/10.1007%2F978-90-481-3132-7_3
- https://www.slideshare.net/pratyayseth/real-time-pcr-34159486
- https://www.thermofisher.com/np/en/home/life-science/pcr/real-time-pcr/real-time-pcr-learning-center/real-time-pcr-basics/essentials-real-time-pcr.html
- https://www.sciencedirect.com/topics/neuroscience/real-time-polymerase-chain-reaction

Great work Sagar,
Keep it up
Thank you very much
Really helpful
best explanation!! looking for further articles!
Very helpful explanation thank you!
Logo T + Hexamer primers both are used
Hexamer primers because of low processivity of reverse transcriptase and oligo T because they bind to oligo A tail of mRNA
This is good explained I inspire it’s
Hi I want to know which primer is used for reverse transcription for rna
Oligo DT and Random hexamer is used
Thank you so much. I really appreciate reading, well explained.
Thanks for details informatio.
Really, it is interesting notes….Every person can understand easily and get very useful information from it. But I want to know protocols should be followed for isolation of RNA from Plant seeds for viral disease identification. Would you tell me the protocols that I should follow for isolation of RNA from plant seeds,,,,?
thanks very very much i got very very important information.
Thanks a lot for your clear Explanation
This is fantastic and vividly illustrated
I am going to apply it in my upcoming research work.
Kudos for a good job done
This is the best explanation I have read. Thank you so much for your time. Your illustrations are self explanatory. Once again, thank you.
Thank you so much for inspiring comment 🙂