Proteins are complex biological molecules composed of amino acids. They are polypeptide structures made up of long chains of amino acid residues. Proteins are one of the most abundant organic molecules that perform diverse functions in living organisms. They act as structural components, catalysts, hormones, enzymes, and regulators of cellular processes. Proteins are also involved in DNA replication, molecule transport, catalyzing metabolic reactions, and providing structural support to cells.
Protein structure is the three-dimensional arrangement of atoms in a protein. There are thousands of proteins in each cell in a living system, each with a unique function and structure. The unique structures of proteins determine their functions. Understanding the structure of proteins is important as it reveals the mechanism involved in various cellular processes.
Interesting Science Videos
Basics of Protein Structure
The basic building blocks of proteins are amino acids. These are organic molecules that contain a central carbon atom, an amino group, a carboxyl group, and a distinctive side chain. These amino acids are bonded together by peptide bonds, forming long protein chains. In this chain, there are two ends: the starting point is called the amino terminus (N-terminus), and the ending point is called the carboxyl terminus (C-terminus). Proteins are built from only twenty amino acids, each with a distinct side chain.
Each amino acid side chain has different properties. The unique properties of the side chains play an important role in shaping the protein’s characteristics. Some side chains are acidic or basic, contributing to the protein’s preference for acidic or basic environments. Others are polar, making the protein soluble in water, while non-polar side chains make the protein soluble in lipids.
Levels of Protein Structure
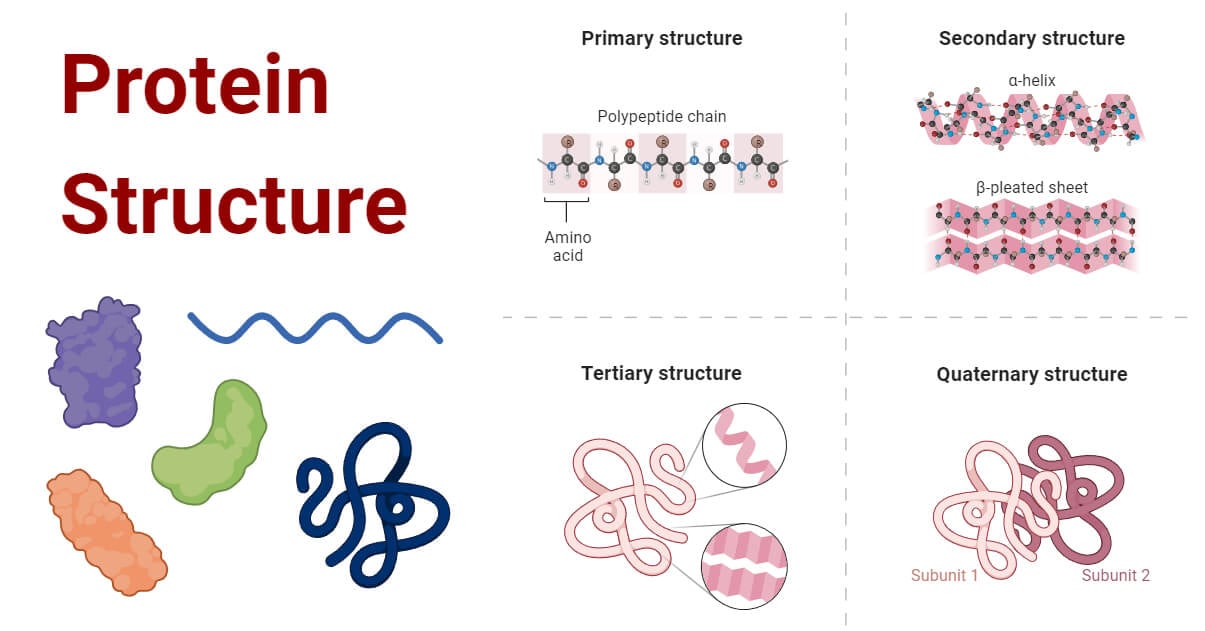
Based on the level of their structure, proteins can be categorized into four structural levels: primary, secondary, tertiary, and quaternary structures.
1. Primary Structure of Protein
- The first level of protein structure is the primary structure which is defined as the linear sequence of amino acids joined together by peptide bonds. The primary structure is maintained by peptide and disulfide bonds.
- The primary structure of a protein is determined by the specific order in which the genetic code is read from the corresponding gene. This genetic information is transcribed from DNA to mRNA and then translated by ribosomes into a sequence of amino acids, forming a peptide chain.
- These polypeptide chains, although initially linear, fold and coil into specific three-dimensional shapes, giving proteins their unique and stable spatial structures.
- The characteristics of specific amino acids in the primary structure influence how the proteins fold at the secondary, tertiary, and quaternary levels.
- Even a small change in one amino acid due to a genetic mutation can lead to significant changes in the entire protein’s structure. For example, in sickle-cell anemia, a single amino acid change can impact the function of hemoglobin.
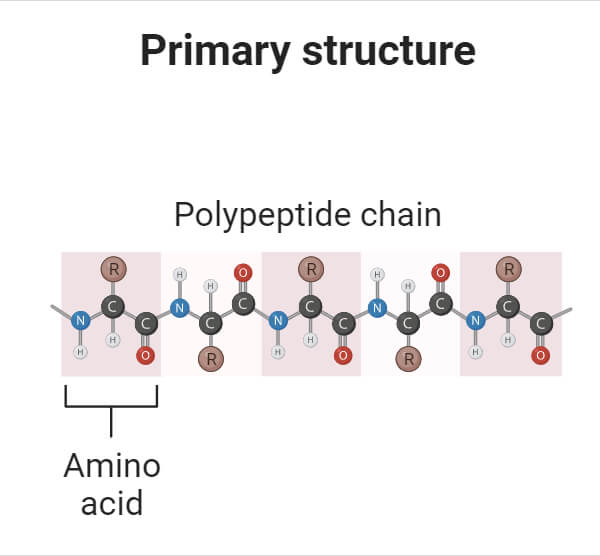
2. Secondary Structure of Protein
- The secondary structure of a protein is the specific spatial arrangement of a particular peptide segment. It refers to the specific way amino acid residues are arranged closely together in the polypeptide chain.
- This arrangement occurs in certain regions where the chain folds locally, forming the secondary structure of the protein.
- This level of structure involves folded or twisted conformation of polypeptides that are stabilized by hydrogen bonds.
The two most common secondary structures of proteins are alpha-helix and beta-pleated sheets.
- α-helix: α helix is a cylindrical structure created when a single chain of amino acids twists around itself. Alpha helixes contain amino acids in a coiled or spiral confirmation. In this helix, a hydrogen bond connects every fourth link in the amino acid chain. This bonding pattern gives rise to a regular helix, completing one full turn every 3.6 amino acids. α helixes are common in proteins found within cell membranes, like transport proteins.
- β-pleated sheet: Beta-pleated sheets are a common structural element in proteins formed by adjacent strands of amino acids, where neighboring polypeptide chains are aligned side by side and connected by hydrogen bonds. These sheets can form in two ways: with adjacent chains running in the same direction (parallel beta sheets) or with chains running in opposite directions (antiparallel beta sheets). In both types, distinct hydrogen-bonding patterns occur, giving rise to the specific structure of beta sheets. Beta sheets contribute to the stable and rigid structure of proteins.
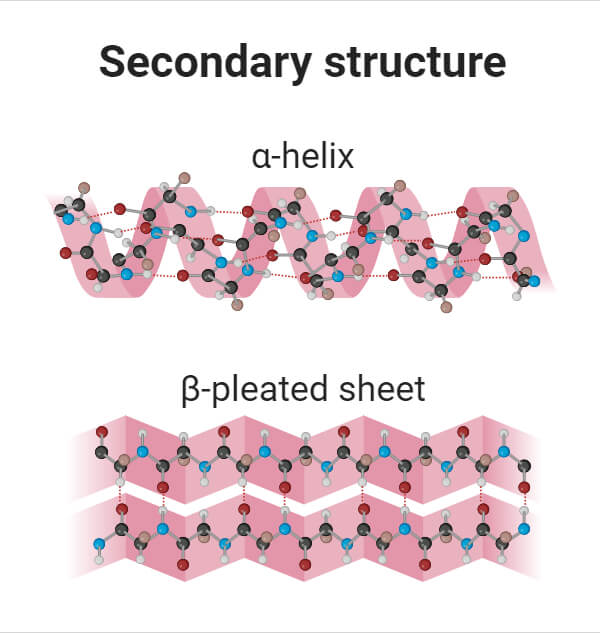
3. Tertiary Structure of Protein
- The tertiary structure of a protein refers to the three-dimensional arrangement of all atoms of a protein formed after the polypeptide chain folds and twists further.
- This arrangement is determined by the interactions of side chains of the polypeptide backbone.
- The tertiary structure of a protein is stabilized by five types of interactions: hydrophobic interaction, ionic interaction, hydrogen bond, Vanderwaal’s interaction, and disulfide bond.
- The tertiary structure also involves the formation of side-chain conformations based on the peptide backbone’s conformation. These conformations create distinct regions known as protein domains. These domains are tightly packed, globular structures within a protein subunit.
- A protein may contain one or several unique domains connected by loop regions. These domains usually contain 100–200 amino acid residues and contribute to both functional as well as structural properties of the protein.
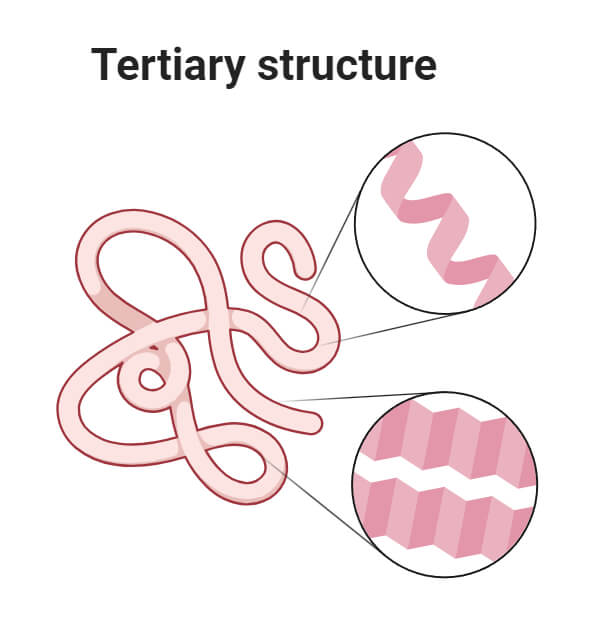
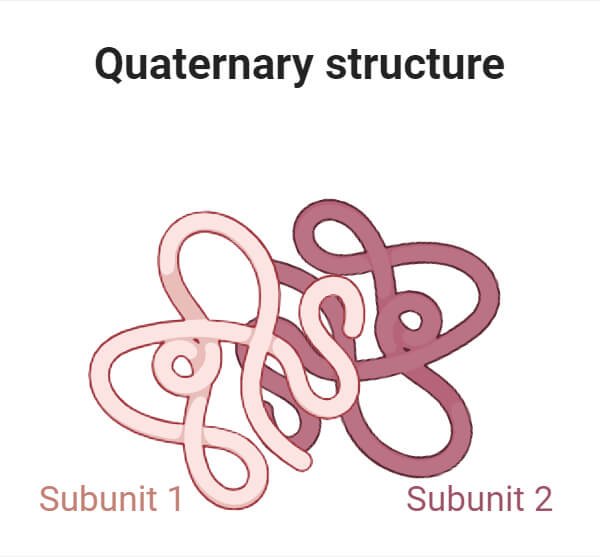
4. Quaternary Structure of Protein
- Proteins containing more than one polypeptide chain exhibit a fourth level of structural organization known as quaternary structure. Each polypeptide chain within such proteins is known as a subunit.
- Quaternary structure is defined as the spatial arrangement of these subunits and their interactions.
- The simplest sort of quaternary structure involves two identical subunits that form a dimer. However, more complex quaternary structures are also common.
- The quaternary structure is formed from the interactions between the side chains of two or more polypeptides and affects the overall shape of the protein.
- Each subunit has its own primary, secondary, and tertiary structures that are held together by hydrogen and van der Waals forces.
- The subunits must be organized precisely for the entire protein to work correctly. Any changes in how the subunits are structured can significantly impact the biological functions of the proteins.
Methods for studying protein structure
Various techniques are used to determine protein structures, such as X-ray crystallography, NMR (Nuclear Magnetic Resonance) spectroscopy, and electron microscopy, each with its advantages and disadvantages.
- X-ray crystallography is one of the commonly used methods to determine protein structures. It involves crystallizing the protein and exposing it to X-rays. The diffracted X-ray pattern is analyzed to create an electron density map, which is then interpreted to locate each atom in the protein. This method provides detailed atomic information about proteins and other molecules in the crystal. However, it works best for rigid proteins forming well-ordered crystals, while flexible proteins are harder to study due to the requirement for precise molecular alignment.
- NMR spectroscopy is another important method used for studying protein structures. It involves placing purified proteins in a magnetic field and probing them with radio waves. The resulting resonances provide information about the conformation of atoms, which is used to create a model of the protein’s structure. NMR is valuable for studying flexible proteins in solution, unlike other methods that require crystallization. Large proteins can present challenges due to overlapping peaks in the NMR spectra.
- Electron microscopy (3DEM) is a technique used to determine 3D structures of large molecules. It involves imaging biomolecules using electron beams and lenses. Cryo-EM, a common method, captures single particles in ice, creating 2D projection images. These images are computationally processed to generate 3D density maps. These maps are interpreted by fitting atomic models into them. Recent advancements in sample preparation, detectors, and data processing have significantly improved 3DEM.
- Experimental data like X-ray diffraction patterns, atomic details from NMR, or overall molecular shapes from electron microscopy alone isn’t always sufficient to create a complete atomic model. Computational tools have also emerged for analyzing protein sequences and predicting structures.
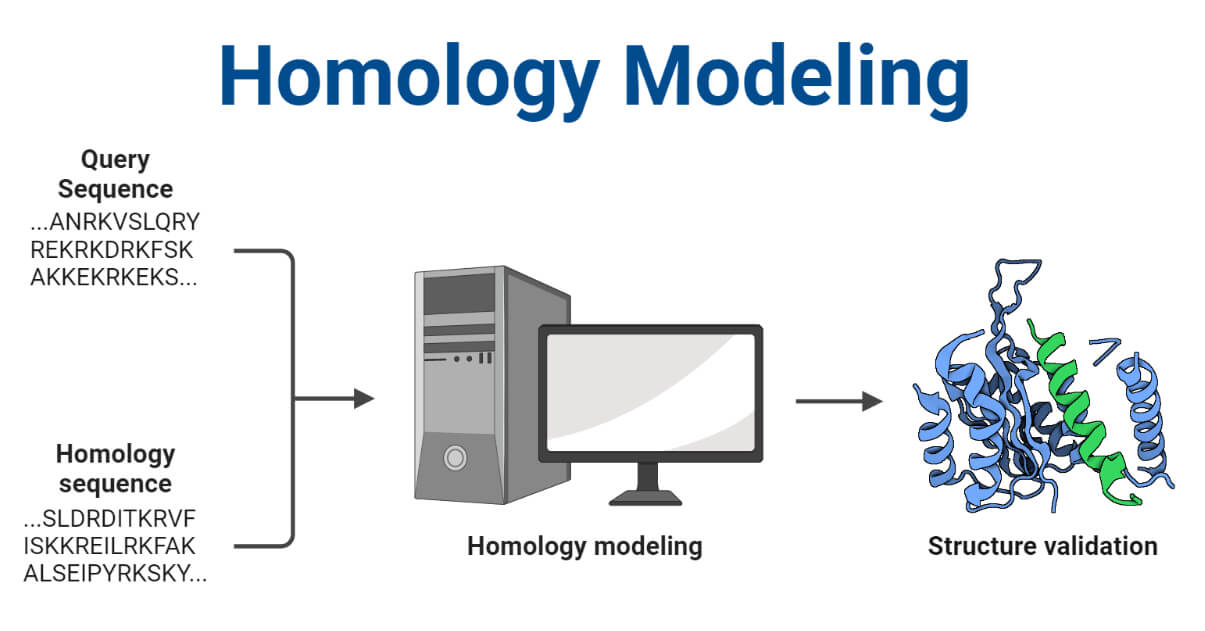
- Homology modeling is one such computational method used to predict the three-dimensional structure of a protein. It relies on a known protein structure (a template) that is closely related to the protein being studied. By comparing the amino acid sequences of the target protein and the template, researchers can create a model of the target protein’s structure.
References
- Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. https://www.ncbi.nlm.nih.gov/books/NBK21054/
- Chapter 2: Protein Structure – Chemistry (wou.edu)
- L. Stryer, (1995) Biochemistry.4th Edition, W. H. Freeman and Company, New York.
- Ouellette, R. J., & Rawn, J. D. (2015). Amino Acids, Peptides, and Proteins. Principles of Organic Chemistry, 371–396. doi:10.1016/b978-0-12-802444-7.00014-8
- PDB-101: Learn: Guide to Understanding PDB Data: Methods for Determining Structure (rcsb.org)
- Protein Structure | Biology for Majors I (lumenlearning.com)
- Protein Structure | Learn Science at Scitable (nature.com)
- Protein Structure | Structure Of Proteins | A-Level Biology Revision Notes (alevelbiology.co.uk)
- Sanvictores, T., & Farci, F. (2022). Biochemistry, Primary Protein Structure. In StatPearls. StatPearls Publishing.
- Skipper, L. (2005). PROTEINS | Overview. Encyclopedia of Analytical Science, 344–352. doi:10.1016/b0-12-369397-7/00493-3
