- Transcription is the process by which the information in a strand of DNA is copied into a new molecule of messenger RNA (mRNA).
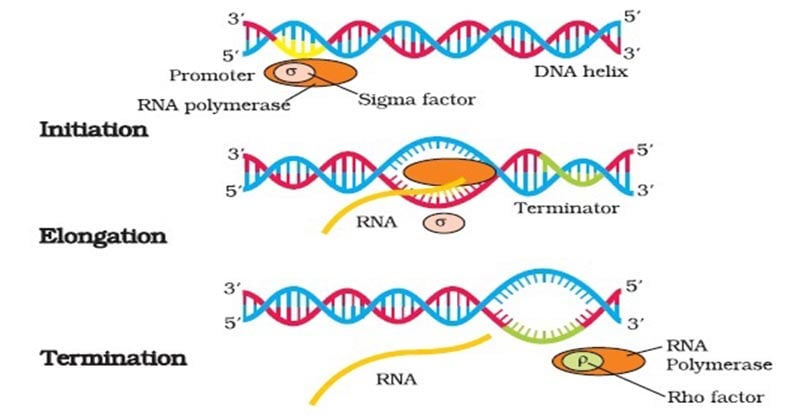
- In prokaryotic organisms transcription occurs in three phases known as initiation, elongation and termination.
Interesting Science Videos
Enzyme(s) Involved
- RNA is synthesized by a single RNA polymerase enzyme which contains multiple polypeptide subunits.
- In E. coli, the RNA polymerase has subunits: two α, one β, one β’ and one ω and σ subunit (α2ββ’ωσ). This complete enzyme is called as the holoenzyme.
- The σ subunit may dissociate from the other subunits to leave a form known as the core enzyme.
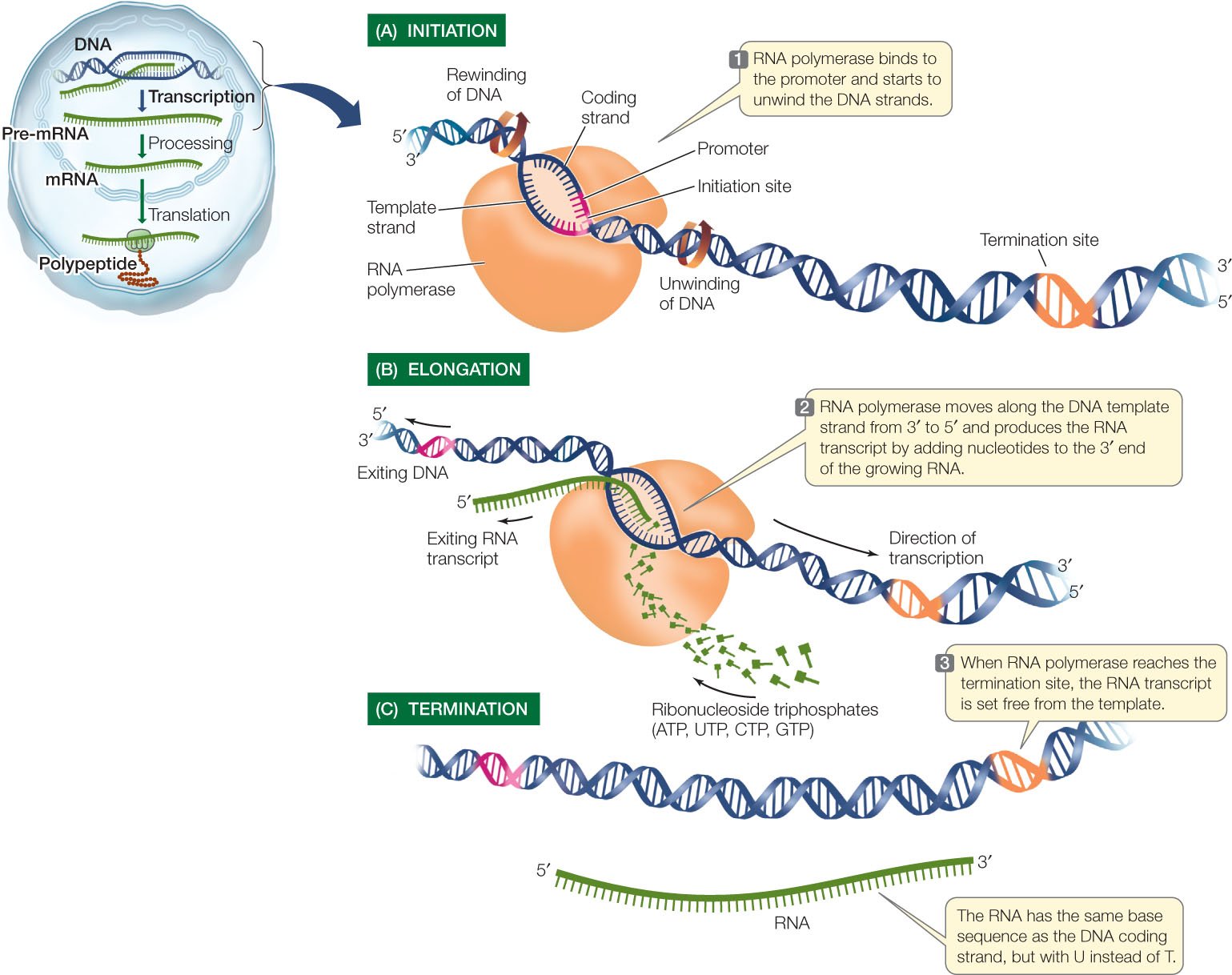
Initiation Phase
During initiation, RNA polymerase recognizes a specific site on the DNA, upstream from the gene that will be transcribed, called a promoter site and then unwinds the DNA locally.
Promoters and Initiation
- The holoenzyme binds to a promoter region about 40–60 bp in size and then initiates transcription a short distance downstream (i.e. 3 to the promoter).
- Within the promoter lie two 6 base pair sequences that are particularly important for promoter function.
- They are highly conserved between species.
- Using the convention of calling the first nucleotide of a transcribed sequence as +1, these two promoter elements lie at positions –10 and –35, that is about 10 and 35 bp, respectively, upstream of where transcription will begin.
- The –10 sequence has the consensus Because this element was discovered by Pribnow, it is also known as the Pribnow box. It is an important recognition site that interacts with the σ factor of RNA polymerase.
- The –35 sequence has the consensus TTGACA and is important in DNA unwinding during transcriptional initiation.
- RNA polymerase does not need a primer to begin transcription; having bound to the promoter site, the RNA polymerase begins transcription directly.
Elongation Phase
- After transcription initiation, the σ factor is released from the transcriptional complex to leave the core enzyme (α2 ββω) which continues elongation of the RNA transcript.
- The core enzyme contains the catalytic site for polymerization, probably within the β subunit.
- The first nucleotide in the RNA transcript is usually pppG or pppA.
- The RNA polymerase then synthesizes RNA in the 5’ →3’ direction, using the four ribonucleoside 5-triphosphates (ATP, CTP, GTP, UTP) as precursors.
- The 3-OH at the end of the growing RNA chain attacks the α phosphate group of the incoming ribonucleoside 5-triphosphate to form a 3’5′ phosphodiester bond.
- The complex of RNA polymerase, DNA template and new RNA transcript is called a ternary complex (i.e. three components) and the region of unwound DNA that is undergoing transcription is called the transcription bubble.
- The RNA transcript forms a transient RNA–DNA hybrid helix with its template strand but then peels away from the DNA as transcription proceeds.
- The DNA is unwound ahead of the transcription bubble and after the transcription complex has passed, the DNA rewinds.
- Thus, during the elongation, the RNA polymerase uses the antisense (-) strand of DNA as template and synthesizes a complementary RNA molecule.
- The RNA produced has the same sequence as the non-template strand, called the sense (+) strand (or coding strand) except that the RNA contains U instead of T.
- At different locations on the bacterial chromosome, sometimes one strand is used as template, sometimes the other, depending on which strand is the coding strand for the gene in question.
- The correct strand to be used as template is identified for the RNA polymerase by the presence of the promoter site.
Termination Phase
- Transcription continues until a termination sequence is reached.
- The most common termination signal is a GC-rich region that is a palindrome, followed by an AT-rich sequence.
- The RNA made from the DNA palindrome is self- complementary and so base pairs internally to form a hairpin structure rich in GC base pairs followed by four or more U residues.
- However, not all termination sites have this hairpin structure. Those that lack such a structure require an additional protein, called rho, to help recognize the termination site and stop transcription.
- Thus the RNA polymerase encounters a termination signal and ceases transcription, releasing the RNA transcript and dissociating from the DNA.
RNA processing
- In prokaryotes, RNA transcribed from protein-coding genes (messenger RNA, mRNA), requires little or no modification prior to translation.
- Many mRNA molecules begin to be translated even before RNA synthesis has finished.
- However, since ribosomal RNA (rRNA) and transfer RNA (tRNA) are synthesized as precursor molecules, they require post-transcriptional processing.
Significance
- Transcription of DNA is the method for regulating gene expression.
- It occurs in preparation for and is necessary for protein translation.
References
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
- Madigan, M. T., Martinko, J. M., Bender, K. S., Buckley, D. H., & Stahl, D. A. (2015). Brock biology of microorganisms (Fourteenth edition.). Boston: Pearson.
- http://www.biologydiscussion.com/rna/transcription/transcription-in-prokaryotes-and-eukaryotes-with-diagram/15546

Awesome notes. Thank you so much Sir.