- DNA replication is the process by which an organism duplicates its DNA into another copy that is passed on to daughter cells.
- Replication occurs before a cell divides to ensure that both cells receive an exact copy of the parent’s genetic material.
- DNA replication uses a semi-conservative method that results in a double-stranded DNA with one parental strand and a new daughter strand.
- Prokaryotic DNA replication is often studied in the model organism coli, but all other prokaryotes show many similarities.
Interesting Science Videos
Features of Prokaryotic DNA Replication
- Replication is bi-directional and originates at a single origin of replication (OriC).
- Takes place in the cell cytoplasm.
- Synthesis occurs only in the 5′to 3′direction.
- Individual strands of DNA are manufactured in different directions, producing a leading and a lagging strand.
- Lagging strands are created by the production of small DNA fragments called Okazaki fragments that are eventually joined together.
Enzymes of DNA Replication
- Helicases: Unwind the DNA helix at the start of replication.
- SSB proteins: Bind to the single strands of unwound DNA to prevent reformation of the DNA helix during replication.
- Primase: Synthesizes the RNA primer needed for the initiation of DNA chain synthesis.
- DNA Polymerase III (DNAP III): Elongates DNA strand by adding deoxyribonucleotides to the 3′end of the chain. Synthesis can only occur in the 5′to 3′direction because of DNAP III.
- DNA Polymerase I (DNAP I): Replaces RNA primer with the appropriate deoxynucleotides.
- DNA topoisomerase I: Relaxes the DNA helix during replication through creation of a nick in one of the DNA strands.
- DNA topoisomerase II: Relieves the strain on the DNA helix during replication by forming supercoils in the helix through the creation of nicks in both strands of DNA.
- DNA ligase: Forms a 3′-5′phosphodiester bond between adjacent fragments of DNA.
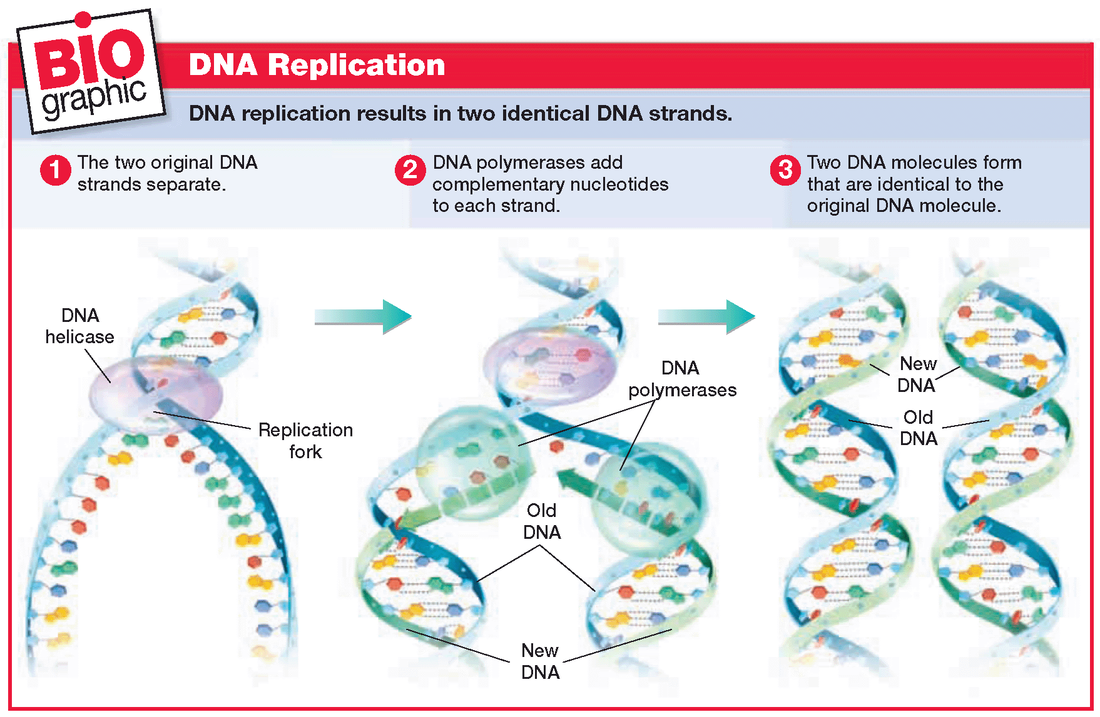
Steps of DNA Replication

- DNA replication begins at a specific spot on the DNA molecule called the origin of replication.
- At the origin, enzymes unwind the double helix making its components accessible for replication.
- The helix is unwound by helicase to form a pair of replication forks.
- The unwound helix is stabilized by SSB proteins and DNA topoisomerases.
- Primase forms RNA primers (10 bases), which serve to initiate synthesis of both the leading and lagging strand.
- The leading strand is synthesized continuously in the 5′to 3′ direction by DNAP III.
- The lagging strand is synthesized discontinuously in the 5′to 3′ direction through the formation of Okazaki fragments.
- DNAP I remove the RNA primers and replace the existing gap with the appropriate deoxynucleotides.
- DNA ligase seals the breaks between the Okazaki fragments as well as around the primers to form continuous strands.
DNA REPAIR
Proofreading:
- In bacteria, all three DNA polymerases (I, II and III) have the ability to proofread, using 3’ → 5’ exonuclease activity.
- When an incorrect base pair is recognized, DNA polymerase reverses its direction by one base pair of DNA and excises the mismatched base.
- Following base excision, the polymerase can re-insert the correct base and replication can continue.
Significance
- DNA replication is a fundamental genetic process that is essential for cell growth and division.
- DNA replication involve the generation of a new molecule of nucleic acid, DNA, crucial for life.
- DNA replication is important for properly regulating the growth and division of cells.
- It conserves the entire genome for the next generation.
References
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
- Madigan, M. T., Martinko, J. M., Bender, K. S., Buckley, D. H., & Stahl, D. A. (2015). Brock biology of microorganisms (Fourteenth edition.). Boston: Pearson.
- https://en.wikipedia.org/wiki/Proofreading_(biology)
- https://sciencing.com/comparing-contrasting-dna-replication-prokaryotes-eukaryotes-13739.html
