Interesting Science Videos
What is chitin?
Chitin is a complex homopolysaccharide consisting of units of amino sugar glucosamine that accounts for the second most abundant polysaccharide of nature after cellulose.
- It is widely distributed in nature, found in the cell walls of fungi, the exoskeleton of arthropods, and certain structures of other invertebrates.
- Chitin, like cellulose, doesn’t accumulate in the biosphere as a result of the extensive hydrolytic activity of soil microorganisms.
- Chitin is associated covalently or non-covalently with other structural molecules as well as the environment.
- The deacetylated derivative of chitin called chitosan has biomedical applications as it acts as an antimicrobial and hydrating agent. The conversion of chitin to chitosan is catalyzed either by enzymatic or chemical hydrolysis.
- The term ‘chitin’ is derived from the Greek word ‘chiton’ which means a coat of mail.
- Depending on the source, chitin occurs in two forms; α and β conformation. A third less discussed γ form is also known.
- These allomorphs differ from one another in their orientation of the micro-fibrils.
- Chitin is considered an essential polymeric structure due to its characteristics like high porosity, biodegradability, predictable degradation rate, and structural integrity.
- Chitin is similar to cellulose in that it is also indigestible by vertebrate animals due to the lack of enzyme system required for its degradation.
Structure of chitin
Chitin is a β(1,4)-linked homopolymer of N-acetyl glucosamine derivative of glucose, and it shares close structural similarity to cellulose.
- In chitin, the alcoholic OH group of the second carbon atom of β-D-glucose units is replaced by an N-acetylamino group.
- Chitin is insoluble in both aqueous and non-polar solvent despite the presence of charges at the acetyl groups.
- Chitin exists as a linear polymer of N-acetyl-D-glucosamine units linked together by β-1,4-glucosidic linkages. This structure results in a three-dimensional α-helix configuration.
- The stability of the α-helix chitin structure is brought about by the hydrogen bonding of the N-acetyl side chains.
- In nature, however, the chitin polymers bind extracellularly by intermolecular hydrogen bonding that forms a crystalline microfibril structure.
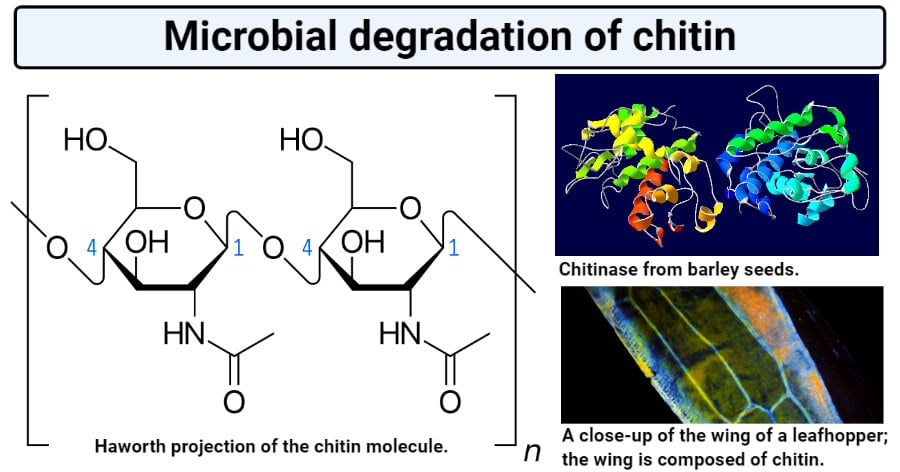
Image Source: Vaccinationist, Jag123, and Zituba.
On the basis of the direction of the chitin fibers and bonds within the polymer, chitin exists in three different conformations.
1. α-chitin
- In α-chitin, the chitin fibers are antiparallel, resulting in an orthorhombic orientation.
- Strong hydrogen bonds stabilize this conformation of chitin structure in the a and b direction while the forces in the c direction are weak.
- Two different types of hydrogen bonding are present in the α-chitin that stabilizes the crystalline structure; intrasheet and intersheet hydrogen bonds.
- The intrasheet hydrogen bond occurs between the carbonyl group of amide I and amide II.
- The intersheet hydrogen bonds exist between the CH2OH side chain and the carbonyl group.
- The decomposition temperature of the crystalline structure of the chitin depends on the hydrogen bonding and thus it is highest at 330°C in α-chitin.
2. β-chitin
- In β-chitin, the chitin fibers are parallel to each other, stabilized only by the intrasheet hydrogen bonding. It consists of monoclinic units.
- The decomposition temperature of the crystalline structure of β-chitin is the lowest at 230°C as it has the lowest number of hydrogen bonds.
3. γ-chitin
- The γ-chitin conformation is characterized by alternating parallel and antiparallel aligned chitin fibers.
- The number and direction of hydrogen bonding in γ-chitin is similar to that in α-chitin. Both intersheet and intrasheet hydrogen bonding exists in γ-chitin.
- The decomposition temperature of the crystalline structure of γ-chitin is 310°C.
What are Chitinases?
- Chitinases are a group of glycosyl hydrolases that range in size from 20kDa to 90kDa and are found in a wide range of organisms like bacteria, fungi, yeasts, plants, actinomycetes, and animals.
- Chitinases degrade chitin directly into smaller, low molecular weight chitooligomers, which serve industrial, agricultural, and medical functions.
- Chitinases works by degrading the β-1,4-linkages that exist between N-acetyl glucosamine units to reduce the length of the polymer, ultimately leading to the formation of monomeric units.
- Chitinases are broadly divided into two groups; exo-chitinases and endo-chitinases.
- Endochitinases split the polymeric chains at random sites internally, thereby forming the dimeric units of di-cetylchitobiose and soluble low molecular mass multimers of glucosamine as chitotriose and chitotetraose.
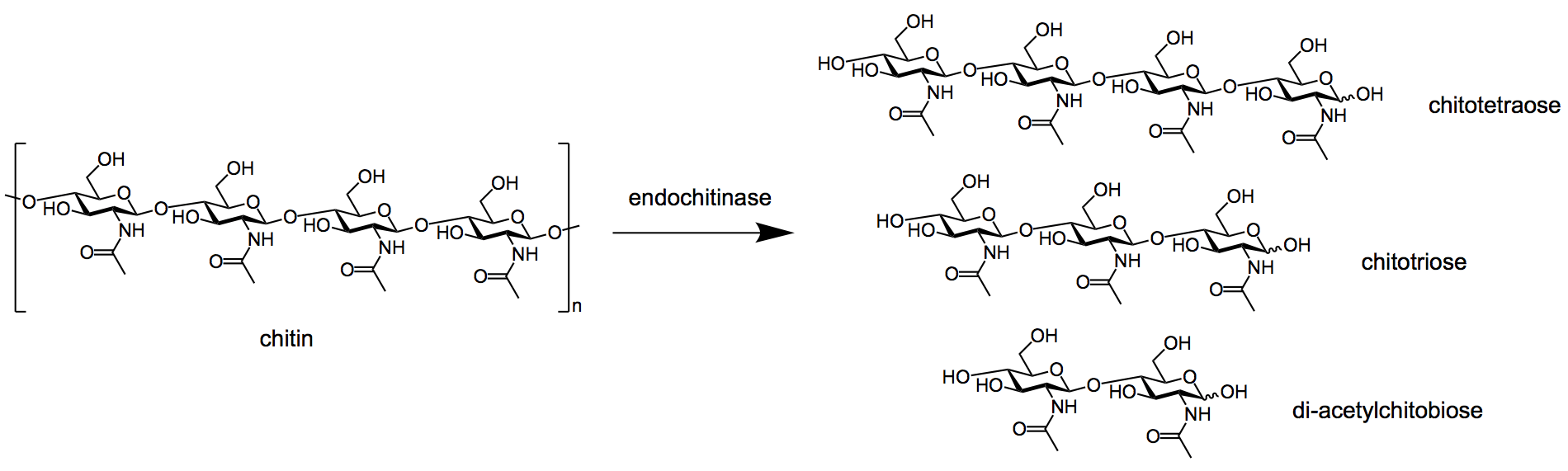
Figure: Endochitinase breaking down chitin into multimer products. Image Source: Wikipedia.
- Exochitinases, in turn, are further divided into two groups; chitobiosidases and 1,4-β-glucosaminidases.
- Chitobiosidases catalyze the subsequent release of di-acetylchitobiose beginning at the non-reducing of the chitin fibrils.
- 1,4-β-glucosaminidases cleave the oligomeric products of endochitinase and chitobiosidases to produce monomeric products of glucosamine.
- Five distinct classes and three families (Family 18, 19, and 20) of chitinases have been proposed based on the amino acid similarity between chitinases from various organisms.
- Family 18 chitinases are mostly sourced from viruses, fungi, bacteria, and some plants. These contain a number of conserved repeats of amino acids and enzyme core with 8 strands of parallel β-sheets.
- Family 19 is of plant chitinases and Treptomyces Family 18 and 19 chitinases do not share amino acid similarity and have a different three-dimensional structure.
- Family 20 contains N-acetylglucosaminidases from bacteria, fungi, and humans.
- Chitinases consist of a diverse group of enzymes that are different in their molecular structure, substrate specificity, and catalytic mechanism.
- Due to their industrial applications, the production of microbial chitinases has increased over the years.
- Methods like fed-batch fermentation, continuous fermentation, and liquid batch fermentation are employed.
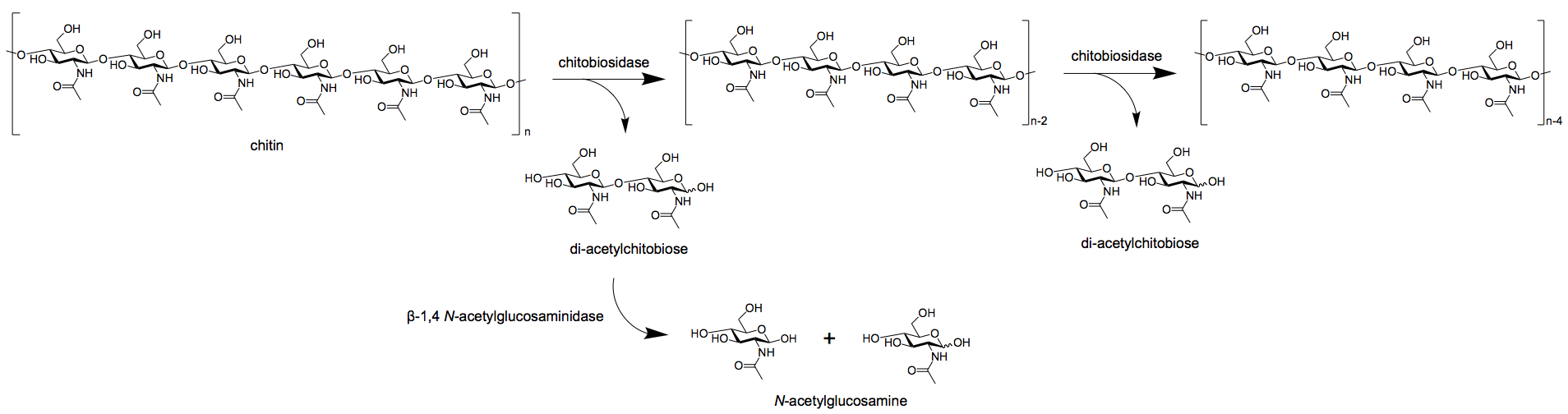
Figure: Exochitinase breaking down chitin into dimers via chitobiosidase and monomers via β-1,4 N-acetylglucosaminidase. Image Source: Wikipedia.
Microorganisms involved in chitin degradation
1. Chitinolytic bacteria
- Chitinolytic bacteria are widely distributed in different habitats, and chitinases are produced by many genera of Gram-negative and Gram-positive bacteria, but not by Archaebacteria.
- Chitinolytic bacteria of the genera Vibrio and Photobacterium are associated with zooplankton and particulate matter that are found in a sea associated with zooplanktons and carapaces.
- Bacterial species of Vibrio, Photobacterium, Aeromonas, Cytophaga, Streptomyces, Photobacterium, Bacillus, Clostridium, and Chromobacterium are well-known chitinolytic bacteria.
- These are specialized utilizers of diacetylchitobiose, and the accumulation of N-acetylglucosamine indicates non-utilizable monomers during random hydrolysis of chitin oligomers.
- Chitinolytic bacteria are also abundant in freshwaters, characteristic genera of Serratia, Chromobacterium, Pseudomonas, Flavobacterium, and Bacillus, with Cytophaga johnsonae.
2. Chitinolytic fungi
- The primary habitat of chitinolytic fungi is the soil where the chitinolytic activity of fungi might even exceed that of bacteria.
- The most common fungal species involved in chitinolysis include Mucorales like Mortierella spp, and Deuteromycetes and Ascomycetes like Aspergillus, Verticillium, Thielavia, Trichoderma, Penicillium, and Humicola.
- The chitinolytic system in these fungi is inducible, and the activity increases with the increase in the chitin-rich substrate.
- Freshwater species Chrytriomyces and Karlingia are obligate chitinophile that degrade chitin to fulfill their nutritional requirement.
3. Slime mold, protozoa, and algae
- Myxomycetes (true slime molds) like Physarum polycephalum are a rich source of lytic enzymes that produce a complex of extracellular chitinases.
- Soil protozoa like Hartmanella and Schizopyrenus, along with slime mold Plasmodium are also known to produce chitinases that participate in the digestion of chitinous food particles engulfed by these invertebrates.
- A colorless heterotrophic diatom, Nitzchia alba, is the only known diatom to digest chitin.
Enzymes involved in the degradation of chitin
There are different classes and families of chitinases that act on different stages of chitin degradation and might even utilize different mechanisms of degradation. There important families of chitinases include family 18, 19, and 20 chitinases. These chitinases might differ in their source and their structural components.
1. Family 18 chitinases
- Family 18 chitinases are retaining enzymes that include both chitinases as well as chitosanases.
- These are found in many organisms, including archaea, bacteria, eukaryote, and viruses. This family of enzymes is widely studied.
- These chitinases are further classified into subclass A, B, and C on the basis of their amino acid sequence similarities.
- In some of the chitinases of the family 18, only a catalytic domain is found, whereas others might have one or more carbohydrate-binding modules.
- The mechanism of catalysis in family 18 chitinases has some modifications compared to the typical double retaining mechanism.
- Instead of using a carboxylate side chain of the enzyme as the catalytic nucleophile, family 18 chitinases use the acetamido group of the C-1 sugar.
2. Family 19 chitinases
- Family 19 chitinases are different from family 18 chitinases as they use an inverting mechanism leading to α-anomeric hydrolysis rather than the retaining mechanism.
- Traditionally, family 19 chitinases were known to exist only in plants, but some bacterial chitinases are also added to the family over the years.
- The family 19 chitinases consist of catalytic domains that have a lysozyme-like fold with shallow substrate-binding grooves that are not rich in aromatic residues.
- Due to the lack of information about the structure of these enzymes, information on their interaction with the substrate is also limited.
3. Chitin deacetylases
- Chitin deacetylases include enzymes like peptidoglycan N-acetyl glucosamine deacetylase and peptidoglycan N-acetylmuramic acid deacetylase that removes the acetyl groups in the substrates.
- These enzymes are essential as they reduce the branching in the structure, which reduces the steric hindrance for other exo and endoenzymes.
Factors affecting chitin degradation
Chitin degradation in soil or on artificial media can be affected by several factors, some of which are:
1. Moisture content
- The process of chitin degradation occurs rapidly in the presence of free water and complete saturation.
- However, the increase in the amount of water has a minimal effect on the degradation process until aeration becomes impaired due to logging.
2. Added glucose
- The addition of glucose in the media or soil decreases the rate of chitin degradation as the organisms tend to utilize the readily available source rather than chitin.
- Glucose is a ready energy source which is easy to metabolize. This, in turn, causes a delay or decreased chitin degradation.
- In the absence of these sources, however, chitin degradation enhances.
3. Aeration
- Since most of the chitinolytic microorganisms are aerobic and thrive in high-oxygen environments, the rate of chitin degradation also increases.
- Some amount of degradation can also be observed in some concentration of CO2 as it allows facultative aerobes and anaerobes to be involved.
- Pure oxygen environment might be toxic in some cases, especially when readily energy source is available.
4. Organic matter
- The presence of organic matter rich in chitin also increases the rate of chitin degradation.
- The increase in organic matter increases the substrate concentration. The rate of degradation might be slow at first as the microorganisms utilize more readily available energy forms, followed by chitin degradation.
- Other forms of energy like cellulose and lignin should also be present as it allows the growth of microorganisms for the formation of proteins and enzymes.
Process (Simple Steps) of chitin degradation
The hydrolysis of chitin occurs in a two-step process;
1. Depolymerization
- Depolymerization is the process of reduction of chitin polymer length by the breakdown of β-1,4 linkages between the N-acetyl glucosamine units.
- This process results in the release of N-acetylglucosamine units by the action of chitinases of chitosanases.
- The initial enzymatic action is of chitinases which are either endochitinases or exochitinases, resulting in the formation of chitobioses or chitotrioses.
- The chitobioses are further acted upon by exoenzymes like chitobioases to form monomeric units.
- In some cases, chitin might be converted into chitosan, which requires the action of chitsonases.
2. Deacetylation
- Depolymerization is followed by acetylation which causes the release of glucosamine units and acetic acid.
- Chitin deacetylases act on the N-acetyl glucosamine dimer or trimers, resulting in the catalytic degradation of the larger molecule into smaller ones.
- The end products of this step are glucosamine and acetic acid, which are then utilized by the microorganism for various purposes.
Mechanisms of microbial degradation of chitin
- The vast amount of chitin produced by different sources is balanced by an equal rate of recycling of the substrate.
- Most of the chitin degradation occurring in nature is microbial, carried out by a different group of microorganisms.
- Chitin degradation occurs in different habitats like the sea, animal guts, and the soil.
- Microbial chitin degradation occurs by one of the two mechanisms; chitinoclastic mechanism and deacetylation mechanism.
1. Chitinoclastic
- Chitinoclastic mechanism of chitin degradation occurs solely by the hydrolysis of glycosidic bonds. Organisms that degrade chitin by this mechanism are called chitinolytic organisms.
- In this mechanism, the substrate is acted upon by the chitinolytic system, consisting of chitinases.
- Exochitinase breakdown acetylchitobiose units from the non-reducing end of the polysaccharide chain.
- Endochitinase cleaves glycosidic linkages randomly along the chain, eventually resulting in the formation of diacetylchitibiose as the major product, along with some tri-acetyl chitotriose.
- The activities of these enzymes may not always be distributed, as the action of these enzymes is dependent on the nature of the substrate.
- Chitobiose (structurally, diacetylchitobiose) is hydrolyzed to N-acetylglucosamine by β-N-acetylglucosaminidase.
- In some cases, β-N-acetylglucosaminidases might also act weakly as exochitinases, cleaving monosaccharide units from the non-reducing ends of the polymeric chains.
- Together, the chitinases and β-N-acetylglucosaminidases form ‘the chitinolytic system’.
2. Deacetylation
- Deacetylation is an alternate mechanism of chitin degradation which involves the conversion of chitin into chitosan.
- This mechanism of chitin degradation is important in the freshwater system or soil sediments.
- The group of enzymes involved in the deacetylation mechanism is termed deacetylases. These enzymes catalyze the process of deacetylation of N-acetylglucosamine polymer.
- The hydrolysis of chitosan occurs in the presence of chitosanases that breakdown the linkages between the β-glucosamine units linked together by β-1,4-glycosidic linkages.
- This cleavage results in the release of chitobiose (glucosaminyl-(1-4)- β-glucosaminide) which is then further degraded by glucosoaminidase to obtain glucosamine units.
References
- Moussian B. (2019) Chitin: Structure, Chemistry and Biology. In: Yang Q., Fukamizo T. (eds) Targeting Chitin-containing Organisms. Advances in Experimental Medicine and Biology, vol 1142. Springer, Singapore. https://doi.org/10.1007/978-981-13-7318-3_2
- Casadidio C, Peregrina DV, Gigliobianco MR, Deng S, Censi R, Di Martino P. Chitin and Chitosans: Characteristics, Eco-Friendly Processes, and Applications in Cosmetic Science. Mar Drugs. 2019;17(6):369. Published 2019 Jun 21. doi:10.3390/md17060369
- Tharanathan RN, Kittur FS. Chitin–the undisputed biomolecule of great potential. Crit Rev Food Sci Nutr. 2003;43(1):61-87. DOI: 10.1080/10408690390826455. PMID: 12587986.
- Lenardon MD, Munro CA, Gow NA. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. 2010;13(4):416-423. doi:10.1016/j.mib.2010.05.002
- Elieh-Ali-Komi D, Hamblin MR. Chitin and Chitosan: Production and Application of Versatile Biomedical Nanomaterials. Int J Adv Res (Indore). 2016;4(3):411-427.
- Hamid R, Khan MA, Ahmad M, et al. Chitinases: An update. J Pharm Bioallied Sci. 2013;5(1):21-29. doi:10.4103/0975-7406.106559
- Abhishek Singh Rathore, Rinkoo D. Gupta, “Chitinases from Bacteria to Human: Properties, Applications, and Future Perspectives”, Enzyme Research, vol. 2015, Article ID 791907, 8 pages, 2015. https://doi.org/10.1155/2015/791907
- Gooday, G.W. Physiology of microbial degradation of chitin and chitosan. Biodegradation1, 177–190 (1990). https://doi.org/10.1007/BF00058835.
Sources
- 1% – https://www.sciencedirect.com/science/article/pii/S0038071708004288
- 1% – https://www.researchgate.net/profile/Malik_Mobeen_Ahmad/publication/236115053_Chitinases_An_update/links/0c96051af02e9c8f3f000000.pdf
- 1% – http://nopr.niscair.res.in/bitstream/123456789/29743/1/IJEB%2052%2811%29%201025-1035.pdf
- <1% – https://www.youtube.com/watch?v=pCowpxUsAN4
- <1% – https://www.sciencedirect.com/topics/chemistry/chitosan
- <1% – https://www.sciencedirect.com/science/article/pii/S0144861710004455
- <1% – https://www.sas.upenn.edu/~caramboc/protein%20lesson.pdf
- <1% – https://www.researchgate.net/publication/8173073_Chitinolytic_activity_of_filamentous_fungi
- <1% – https://www.researchgate.net/publication/261031080_Hydrogenases
- <1% – https://www.researchgate.net/publication/236115053_Chitinases_An_update
- <1% – https://www.ncbi.nlm.nih.gov/pubmed/26542048
- <1% – https://www.ncbi.nlm.nih.gov/pubmed/24095741
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6097792/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4926359/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3551169/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2851747/
- <1% – https://www.mdpi.com/1422-0067/19/2/412/pdf-vor
- <1% – https://ui.adsabs.harvard.edu/abs/2014BGeo…11.3339W/abstract
- <1% – https://pubs.acs.org/doi/10.1021/acs.chemmater.8b05183
- <1% – https://portlandpress.com/bioscirep/article/38/4/BSR2018032300/58006/Chitinase-diversity-limitations-and-trends-in
- <1% – https://en.wikipedia.org/wiki/Chitin
- <1% – https://edubuzznotes.com/hydrogen-bond/
- <1% – http://europepmc.org/articles/PMC5094803
