The genetic composition of an individual cell or organism that determines or influences its phenotype is known as its genotype.
- From embryonic development to adulthood, a person’s genotype defines their inherited potential and limitations.
- The dominant relationships of the alleles that make up an individual’s genotype, which consists of the full complex of genes inherited from both parents and environmental factors, define the actual appearance and behavior of the individual; phenotype.
The following relationship has frequently been used to illustrate how components interact:
Genotype + Environment + Random variation → Phenotype
Interesting Science Videos
Genotype Etymology
- The word “genotype” is adapted from the German Genotypus based on the Greek word genea meaning “generation or race.”
- Wilhelm Ludvig Johannsen developed the genotype-phenotype concept in 1909 to distinguish between hereditary and environmental variations.
- He stated that a person’s phenotype represents the observable structural and functional characteristics formed by the interaction between genotype and environment. In contrast, an individual’s genotype represents the totality of hereditary factors.
- This distinction resulted from Johannsen’s research on heritable plant variation, which also impacted his pure-line theory of heredity.
Genes, Alleles, and Genotype
- Genes typically come in pairs, or alleles, in eukaryotes.
- A variant of a particular gene is referred to as an allele.
- The paired alleles will be located at the same location on the chromosome.
- Each parent contributes one of the two alleles for a particular gene since most eukaryotes reproduce sexually.
- Because each parent comes from a different lineage of organisms, random replication errors and environmental interactions with the DNA have uniquely altered each parent’s genes.
- One of the alleles will be expressed while the other is suppressed if the trait inherits in a simple Mendelian pattern.
- The dominant allele is the one that expresses itself, whereas the recessive allele is the one that does not.
- Three distinct genotypes are possible if the dominant allele is “T” and the recessive allele is “t.”. They are “TT,” “tt,” and “Tt.”
- Because the alleles in the pair are identical, i.e., both dominant or both recessive, the term “homozygous” is used to describe the pairs and is represented by “TT” and “tt.” On the other hand, the allelic pair “tt” is referred to as “heterozygous.”
- Individuals having genotypes TT or Tt will display the dominant trait in their phenotypes, whereas those with genotype tt will display the recessive trait.
- Not all traits are determined by a single gene, many genes have multiple allelic variants, and not all gene pairs have a simple dominant/recessive relationship.
- Non-Mendelian inheritance patterns include codominance, partial dominance, and polygenic inheritance.
- Many of the observable traits in humans are non-Mendelian.
- To fully write out an organismal genotype would require a set of characters representing the alleles for each of the 20,000 or so known genes in the human genome.
Punnett square and Genotype
- The Punnett square is an essential genetic tool to predict inheritance patterns and ratios.
- It is a table that lists every potential result of a genetic cross between two individuals with known genotypes.
- It consists of a square divided into four quadrants.
- Humans are diploid organisms.
- One genotype is provided at the top of each column and down the left side of the square for each conceivable genotype of the haploid female gametes.
- One genotype per row is presented for each conceivable genotype of the haploid male gametes.
- The table’s squares, which correspond to the cross’s possible outcomes, can then be filled up.
- The male gamete from the corresponding row and female gamete from the corresponding column combine at fertilization to form the diploid genotype in each square.
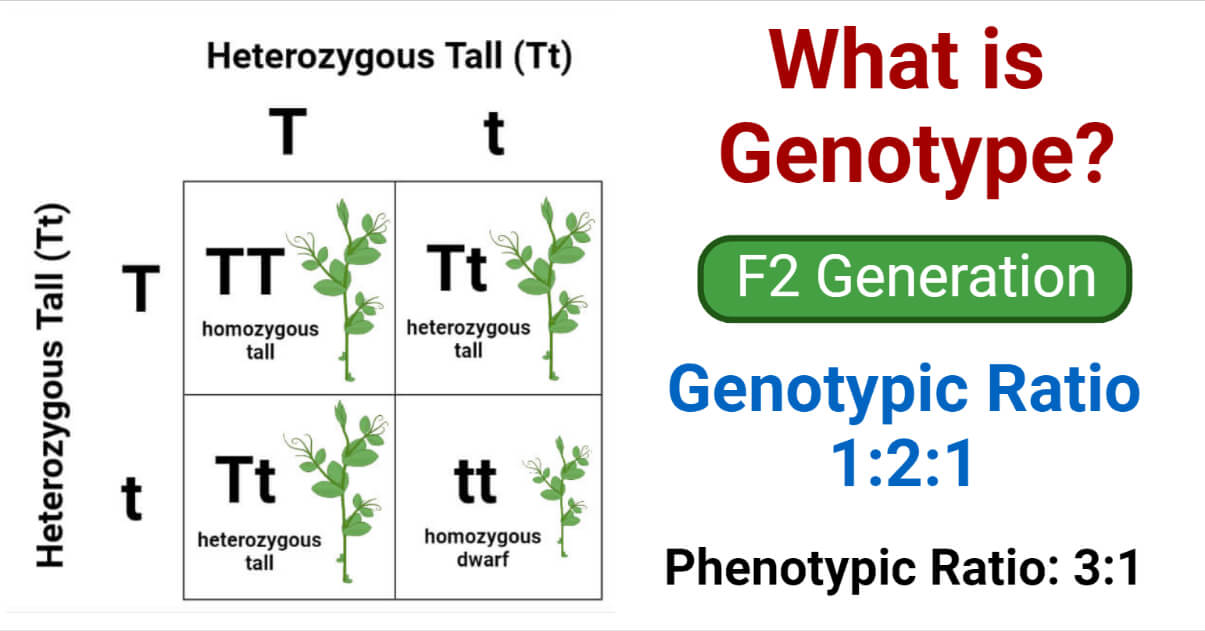
In a monohybrid cross TT x tt, where
- TT – Pure tall plant
- tt – Pure dwarf plant
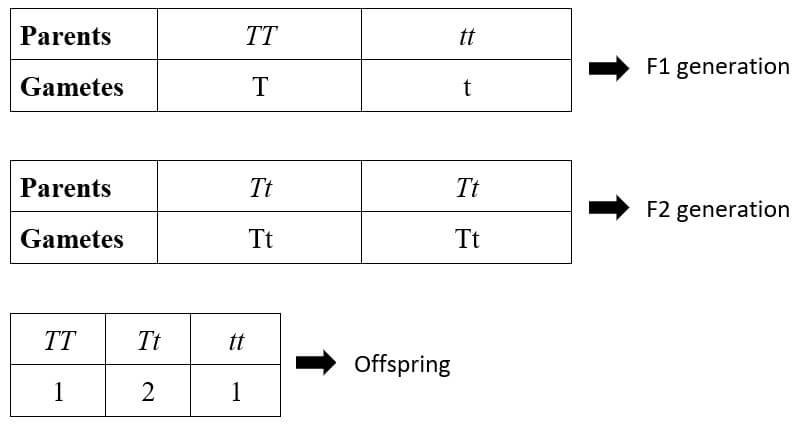
This gives us the 3:1 phenotypic ratio and 1:2:1 genotypic ratio, as described by Mendel.
- TT (Pure tall)
- Tt (Hybrid tall)
- Tt (Hybrid tall)
- tt (Pure dwarf)
Two of the four offsprings, one tall (TT) and one dwarf (tt), are homozygous, while the other two (Tt), both of which express the dominant tall trait, are heterozygous.
Genotyping
- Genotyping determines the DNA sequence or genotype at specific positions within an individual’s genome.
- In linkage and association studies, sequence variants can be employed as markers to find the genes responsible for particular traits or diseases.
- Genotyping is an experimental technique that identifies variations in DNA sequences between people or populations.
- Compared to a reference sequence generated from the general population or a specific subgroup, an individual genome is recognized as a different variant.
- There are many ways in which a variant sequence can diverge from the reference sequence.
- Single nucleotide variations (SNV), single nucleotide polymorphisms (SNPs), insertions and deletions (indels), and copy number variation (CNV) are different types of genetic variation.
- SNPs are the most common sequence variant investigated by researchers; they are typically defined as SNVs that occur at >1% in the population.
- Based on a haploid genome size of 3.1 x 109 bp and the number of SNPs listed in Build 151 of the SNP database, dbSNP, maintained by the National Center for Biotechnology Information (NCBI) (RefSNP count 660,773,127), the human genome should have an SNP around every 9.5 bases.
The frequency of SNPs is also high in other popular model species.
Genotype Network
- In systems biology, genotype networks are used to investigate the “evolvability” or “innovability” of a group of genotypes sharing a single, broadly defined phenotype and to determine if a certain phenotype is resistant to mutations.
- They have been used to determine how much a metabolic network can be adjusted without losing the ability to survive utilizing a certain carbon source to research the evolvability of metabolic networks in simple organisms.
- They have also been applied to research how a metabolic network might “evolve” a new phenotype, such as the capacity to survive on a different carbon source.
- Genotype networks have also been used to explore the stability of RNA folds and protein structures to determine how many mutations can accumulate in a sequence without losing the secondary structure.
References
- Britannica, T. Editors of Encyclopaedia. (2022). genotype. Accessed from: https://www.britannica.com/science/genotype
- Biology Online. (2022). Genotype. Accessed from: https://www.biologyonline.com/dictionary/genotype
- Online Etymology Dictionary. (2022). Genotype. Accessed from: https://www.etymonline.com/word/genotype
- Dall’Olio GM, Bertranpetit J, Wagner A, & Laayouni H. (2014). Human Genome Variation and the Concept of Genotype Networks. PLOS ONE. 9(6): e99424. https://doi.org/10.1371/journal.pone.0099424
- IDT: Integrated DNA Technologies. (2022). Genotyping. Accessed from: https://sg.idtdna.com/pages/applications/genotyping
- J. Phelan. (2013). Punnett Square. In Brenner’s Encyclopedia of Genetics. Second Edition. Academic Press, page 532. ISBN 9780080961569. https://doi.org/10.1016/B978-0-12-374984-0.01242-0.
- Passarge E. (2001). Phenotype and Genotype. In Color Atlas of Genetics. Second Edition. Thieme Stuttgart. New York, pg. 138
- Scitable. (2014). Genotype. Accessed from: https://www.nature.com/scitable/definition/genotype-234/
- Verma P.S. and Agarwal V.K. (2005). Genetics, Human Genetics and Eugenics: Introduction. In Cell Biology, Genetics, Molecular Biology, Evolution and Ecology. Multi-color Edition. S. Chand & Company Ltd. Ram nagar, New Delhi, pg 3-9.
