Exon is defined as the segment of the eukaryotic gene that encodes a portion of the final product of the gene (protein).
Exon = Expressed regions
Exon is also defined as the segment of RNA which remains after the post-transcriptional modification and which is transcribed into protein or incorporated into RNA structure- this part of the gene codes for proteins.
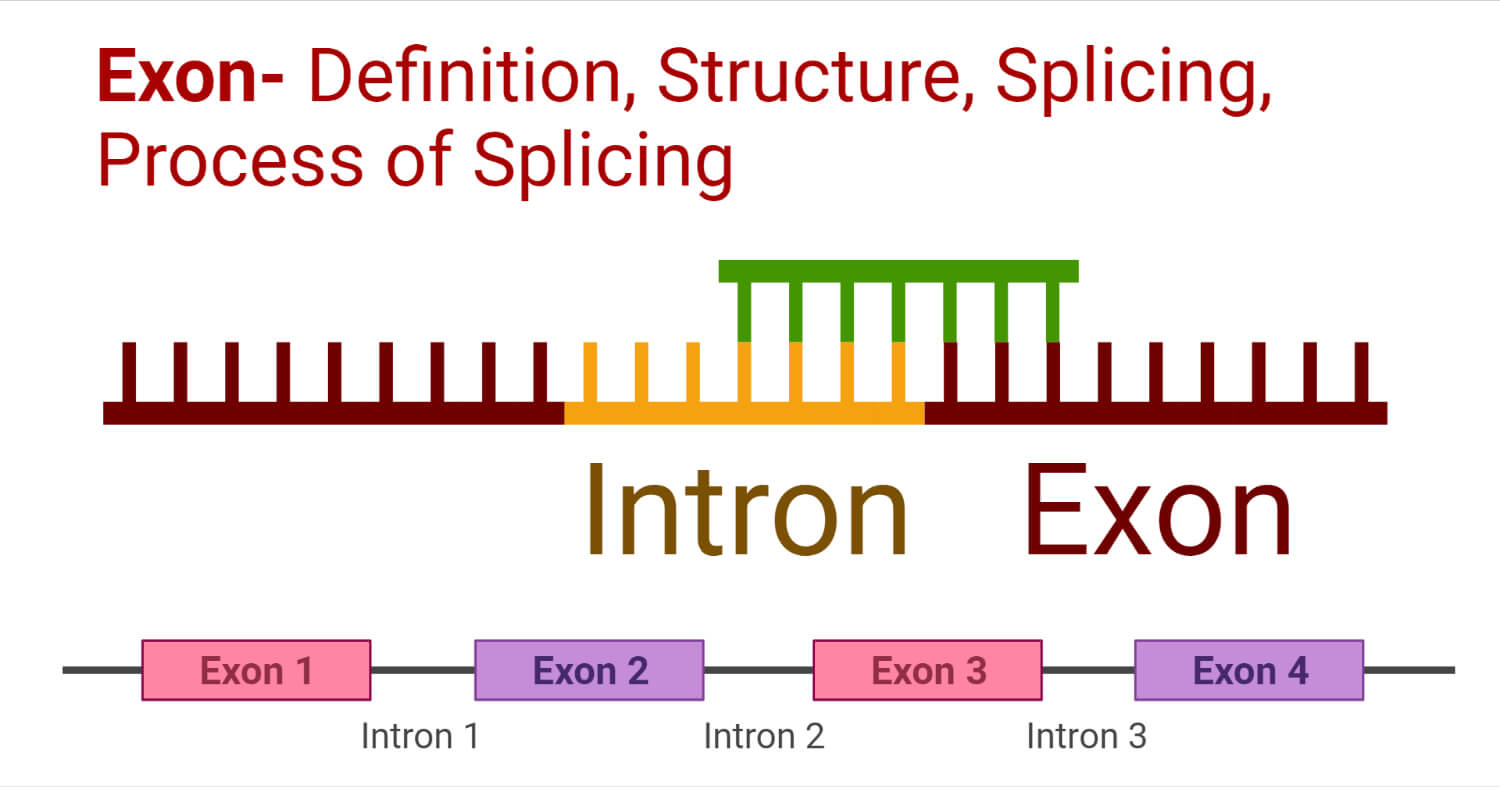
- Exons are exclusively present only in the eukaryotic gene.
- Exons are the coding regions of a gene.
- They code for proteins and are very important segments of the gene.
- Exons are interspaced by introns in a gene.
- The process of splicing (a post-transcriptional process) removes the introns (non-coding regions) and joins the exons.
Interesting Science Videos
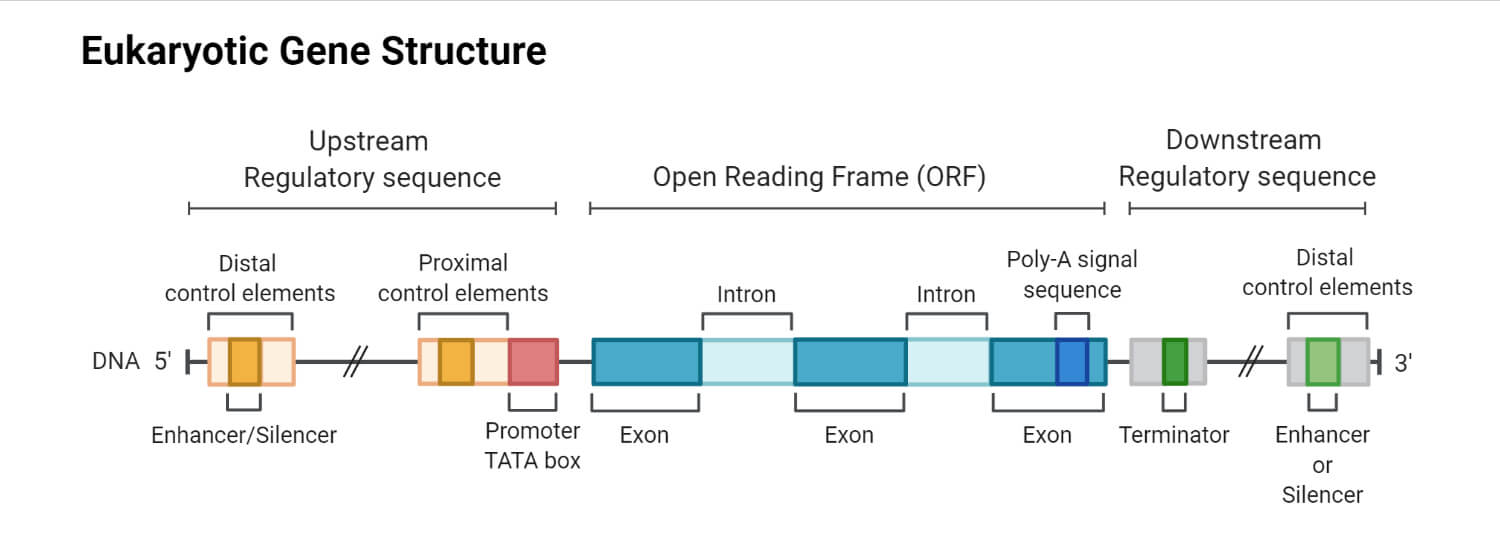
Exon Structure
- Nucleotide sequences in DNA or RNA which are expressed are known as exons.
- Exons are conserved in DNA or mature RNA.
- Exon structure includes the untranslated regions from both 5` and 3`. This includes the start and stops codons and other codons which code for proteins.
Splicing
Splicing is defined as the process in which introns, the non-coding regions, are excised out of the primary mRNA, and exons are joined together in the primary transcript.
- In this process, a pre-mRNA is formed into a mature mRNA.
- Splicing occurs before the process of translation, before protein synthesis.
- It is an important process because correct protein cannot be coded without splicing.
- It also plays an important role in the regulation of gene expression and proteins.
- Several methods for splicing are known, but the process mainly depends on three main factors-
- Type of organism
- Structure of RNA or intron
- Presence of a catalyst
Process of splicing
- The exon-intron junctions in the pre-mRNA have conserved sequences. The 5` end junction of the pre-mRNA has a GU sequence and the 3` end of the junction has an AG sequence.
- These specific sites are known as 5` splice sites and 3` splice sites respectively.
- Along with these two sites, an invariant site is also present with 15-45 nucleotides upstream of the 3` slice site. This site is rich in A bases and is known as Branch point or Branch site.
- Spliceosome recognizes these conserved sequences in the pre-mRNA at the respective splice sites.
- A spliceosome is a set of RNA-protein complexes, and each of these complexes is made up of small nuclear RNA and a protein. Collectively this small nuclear RNA and protein are known as small nuclear ribonucleotideproteins (snRNP).
- The important snRNPs which make up the spliceosome are U1, U2, U5, and U4-U6. (U4 and U6 are bounded together.)
Splicing Pathway
- The first step of splicing involves U1 and U2. U1 binds to the 5` splice site and U2 binds to the invariant site or the branch point.
- In the second step the remaining snRNPs bind to the earlier bound snRNPs. U5 and U4-U6 bind to the intron region and hence, now the spliceosome is assembled.
- Now, the spliceosome loops out the intron, and the two ends of the introns are brought close to each other.
- Further U1 and U4 are released and U6 is bound to both, the 5` splice site and U2.
- In the next step the 5` end of the intron is cleaved and it attaches to the branch point of the intron, which is rich in A.
- Finally, the 3` end of the introns is also cleaved and the intron is released and further degraded by enzymes. The intron structure is called a Lariat (loop-like).
- Lastly the two exons are joined together.
- The snRNPs are used for splicing of other introns and the process of splicing continues for the remaining introns in the pre-mRNA.
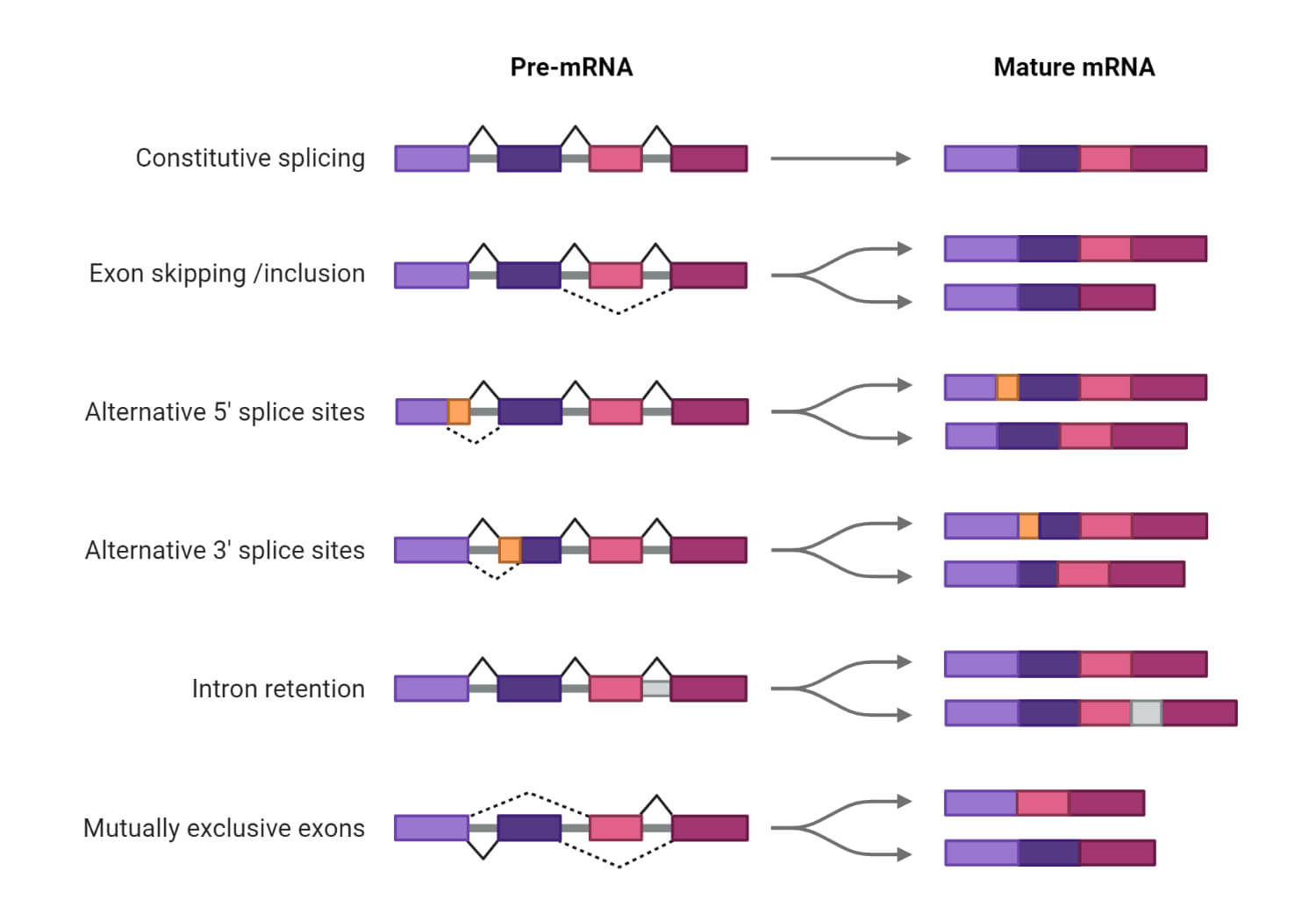
Alternative Splicing
- Alternative splicing is the process in which different variations in the mRNA are created by joining different exons.
- Alternative splicing leads to isoforms of proteins.
- This leads to changes in the chemical and biological activity of proteins.
- This means one gene can code for more than one type of mRNA, and more than one type of protein.
The most common types of alternative splicing are:
- Consecutive splicing – in this splicing process consecutive introns are spliced and consecutive exons are joined together.
- Exon skipping – in this process certain exons along with their adjacent exons are excised from the pre-mRNA before translation.
- Alternative 5` splice site or 3` splice site – this can be achieved by joining of exons t alternative 3` or ` splice site.
- Intron retention – this is achieved when some introns are retained in the mature mRNA.
Self-Splicing Introns
Self-splicing is the process in which the introns (or the RNA) can excise themselves from the pre-mRNA without any precursors and proteins.
It occurs by the mechanism of phosphoester transfer.
Conclusion
- Exons are the coding regions that code for specific amino acids.
- Exons are very important in protein formation.
- Exons are separated by non-coding regions called introns.
- Removal of introns is achieved by the process of splicing.
- Splicing involves the use of cellular machinery known as Spliceosome.
- Due to splicing, the exons are joined together forming a complete gene with all coding regions.
- At the end of the splicing process, a pre-mRNA is converted into a mature mRNA.
- Alternative splicing forms protein isoforms which lead to variations in the biological and chemical activity of proteins. Hence one gene gives rise to more than one type of mRNA.
- Some RNA molecules can undergo self-splicing.
- Splicing is an extremely important process in eukaryotes before translation.
References
- https://www.news-medical.net/life-sciences/What-are-introns-and-exons.aspx
- Gene VIII, Benzamin Lewis
- Lehninger Principles of Biochemistry – 6th ed- c2013-
- https://www.cancer.gov/publications/dictionaries/genetics-dictionary/def/splicing
- https://www.yourgenome.org/facts/what-is-rna-splicing
- https://www.technologynetworks.com/genomics/articles/alternative-splicing-importance-and-definition-351813
- Clancy, S. (2008) RNA splicing: introns, exons, and spliceosome. Nature Education 1(1):31
- https://www.nature.com/scitable/topicpage/rna-splicing-introns-exons-and-spliceosome-12375/
