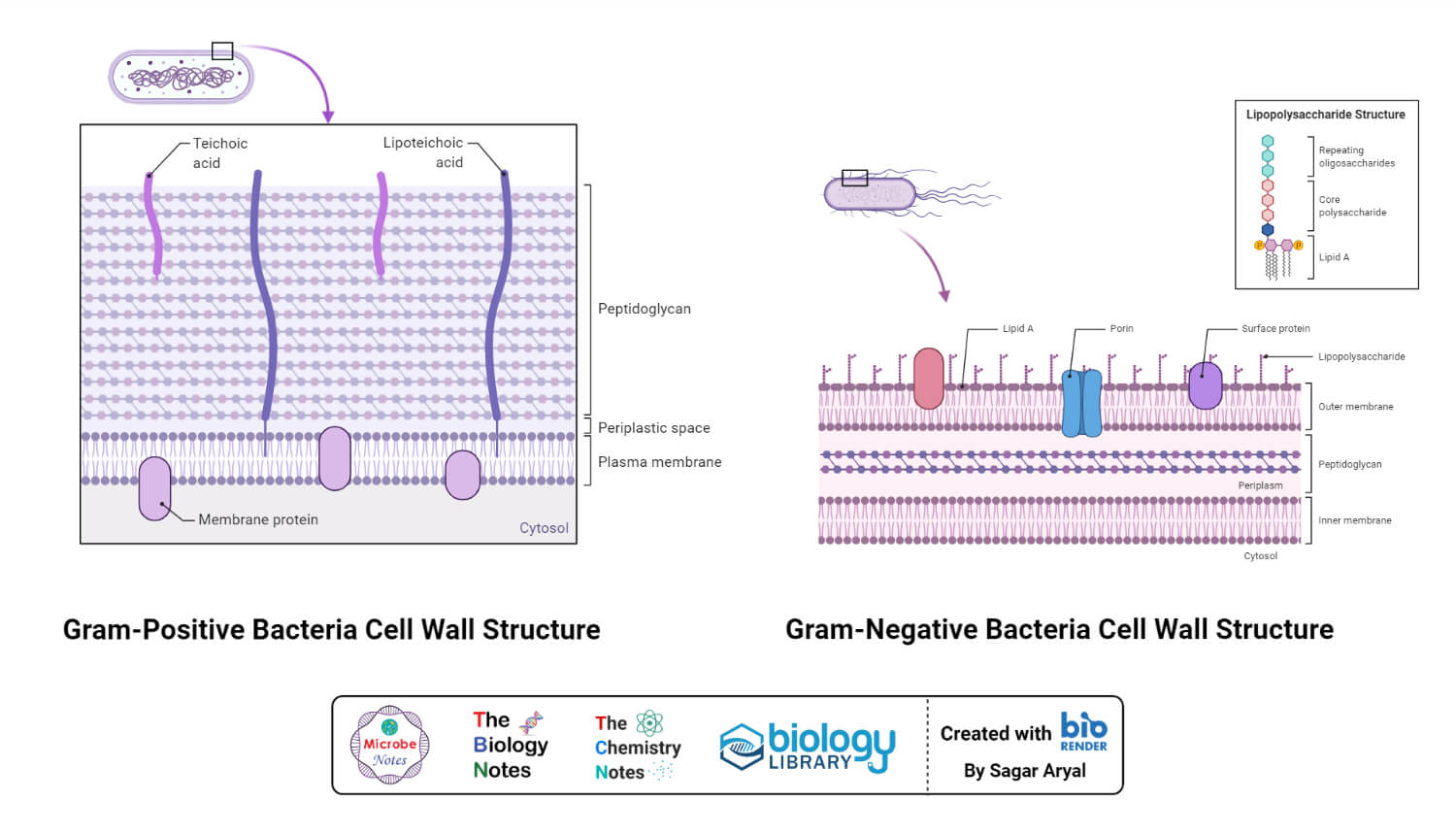
Generally, the bacterial cell consists of a cell wall, cell membrane, and nucleus. The cell wall is the outer covering of the bacteria-containing peptidoglycan layer which is made up of cross-linked polymers. Peptidoglycan is mainly responsible for all the mechanisms including resistivity, virulence factors including- the shape of the bacteria.
Interesting Science Videos
What are Cell wall synthesis inhibitors?
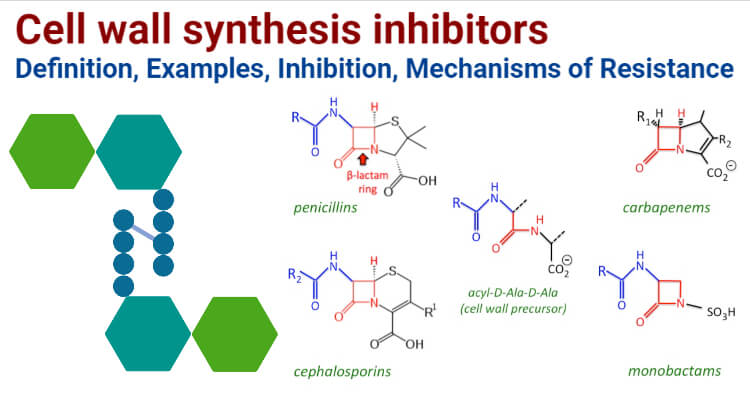
Cell wall synthesis inhibitors include antibiotics of class β- lactams and Glycopeptides. Β- lactam antibiotics consist of Penicillins, Cephalosporins, Monobactams, and Carbapenem. Glycopeptides include Vancomycin and Teicoplanin which are the most commonly used antibiotics of this class. Penicillin includes Ampicillin, Oxacillin, Cephalosporins include Cefpodoxime, Cefepime, Monobactams include Aztreonam which is the only commercially available antimicrobial of this class. Carbapenem consists of Imipenem, Meropenem and Ertapenem. Cell wall synthesis inhibitors are the most used antibiotics for treating Gram-negative as well as Gram-positive infections.
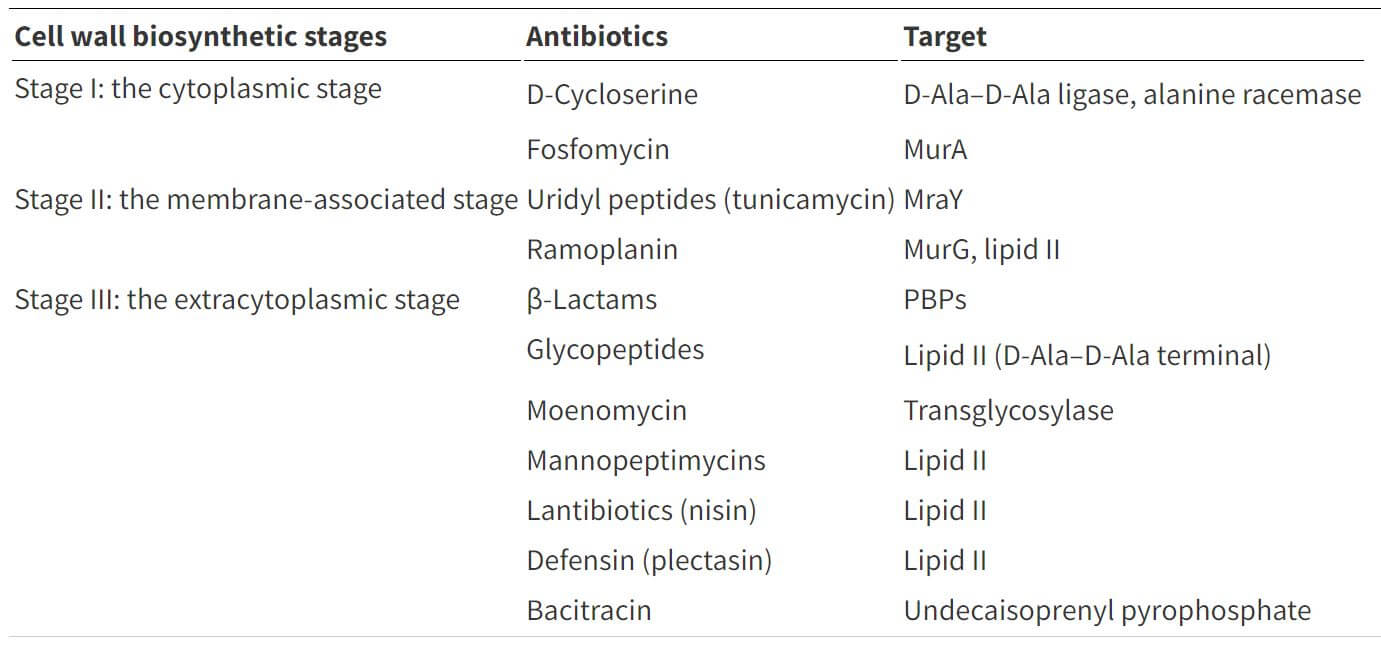
Table: Cell wall biosynthesis inhibitors and their targets. Table Source: https://doi.org/10.1039/C6MD00585C
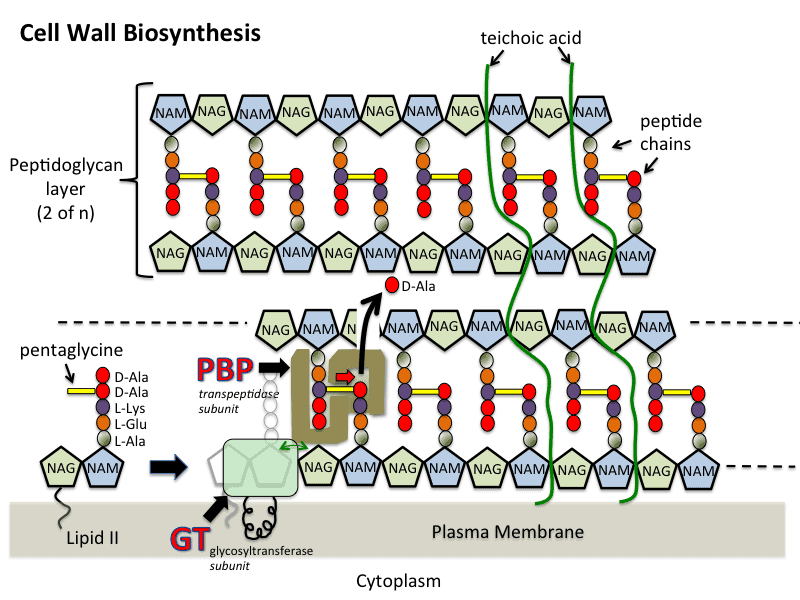
- Peptidoglycan is made up of a polymer of peptides and glycan. Mainly synthesis of peptidoglycan occurs in three stages which include; cytoplasm, membrane, and periplasm.
- In the cytoplasmic stage, UDP-GLcNAc is catalyzed in four steps process. MurA and MurB catalyze forming UDP-MurNAc. Four enzymes Mur C, Mur D, Mur E, and Mur F catalyzes alanine, glutamic acid which are involved in important steps. Alanine ligase and Alanine racemase are two important enzymes involved in the formation of D-Ala-D-Ala.
- The second membrane-associated stage includes the formation of lipid intermediates.
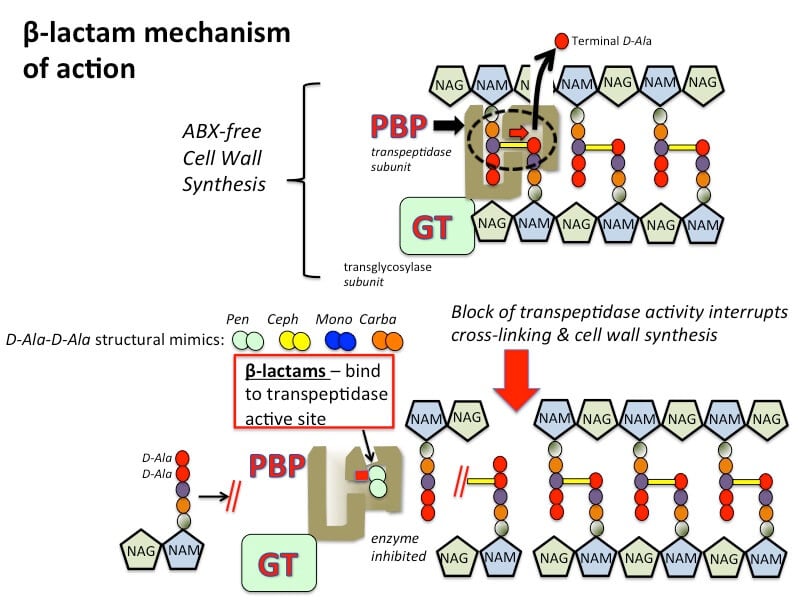
- In the third stage, peptidoglycan chains were formed by glycosyltransferases which are then cross-linked by the enzyme transpeptidases. This enzyme transpeptidase consists of PBPs (Penicillin Binding Proteins) and Rod A
- Mur enzymes, Transpeptidases, glycosylases, and lipid components are the main components involved in cell wall synthesis.
Examples of Cell wall synthesis inhibitors
Beta-lactam antibiotics
a. Penicillins- Ampicillin, Amoxicillin, Piperacillin, Oxacillin
b. Cephalosporins:
- First-generation: Cefazolin, Cefalexin
- Second-generation: Cefoxitin, Cefuroxime
- Third-generation: Cefixime, Ceftazidime, Ceftriaxione
- Fourth-generation: Cefepime
c. Monobactams: Aztreonam, the only commercially available monobactam.
d. Carbapenems: Imipenem, Meropenem
Glycopeptides
Vancomycin, Oritavancin, Teicoplanin, Telavancin, Bleomycin, Ramoplanin, Decaplanin.
Process/Steps of Inhibition
- Beta-lactams interfere in synthesis by acting as a component of D-Alanine- D- Alanine with the help of transpeptidase enzyme in transpeptidase reaction. Glycine residues cross-link amino acid portion of peptide-chain in the presence of penicillin-binding proteins (PBPs). New peptidoglycan chains are formed resulting in cell rupture due to osmotic lysis.
- Bacterial cells contain autolytic enzymes which are known for cell growth and division and beta-lactam antibiotics affect the autolytic activity resulting in bacterial cell death.
- Glycopeptide antibiotics bind to lipid II component that prevents the recycling of central lipid transporter that plays important role in the mode of action. They inhibit peptidoglycan synthesis by linking with the pentapeptide chains and prevent the addition of new peptidoglycan units. Transglycosylation and transpeptidation are inhibited by this interaction resulting in bacterial cell death.
- Vancomycin, a last resort drug of glycopeptides, prevents the binding of D-alanyl with penicillin-binding proteins which inhibits cell wall synthesis causing bacterial lysis.
- Permeabilization and depolarization of bacterial cell membrane resulting in inhibition of cell wall synthesis by some of the glycopeptides.
Mechanisms of Resistance
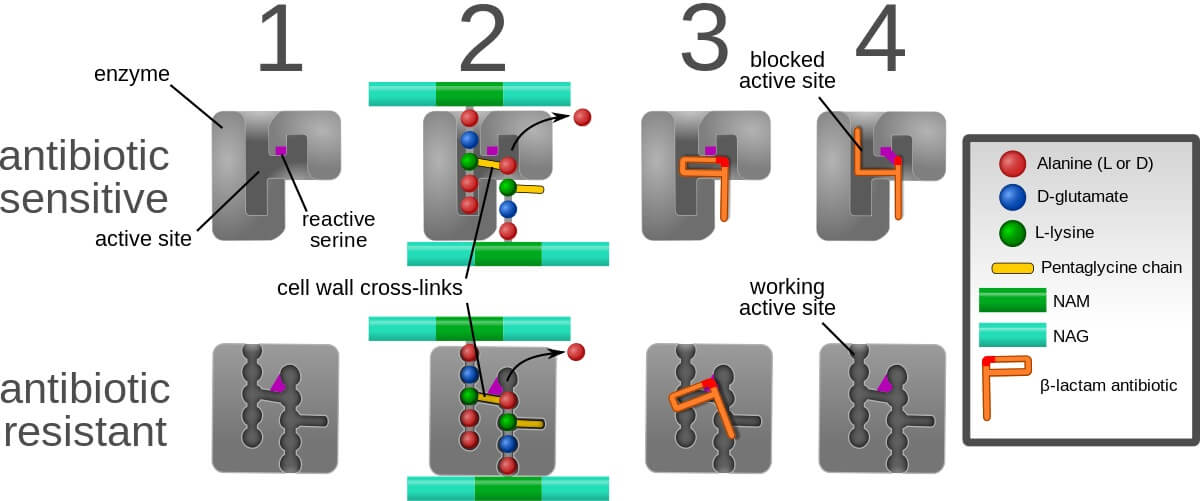
The affinity of the drug, its permeability, and stability determines the activity against bacteria. During the past two decades, resistant organisms towards antibiotics are increasing which is of great concern. Organisms develop resistance to beta-lactam antibiotics through a synthesis of PBP2a (penicillin-binding protein 2a). It has a low affinity allowing transpeptidase activity which results in bacterial colonization in patients. Chromosomal mutations also directly or indirectly increase the level of penicillin-binding protein 2a and transcription causes bacterial resistance. Β-lactams resistance is mainly due to the alteration of Penicillin-binding proteins and enzymatic degradation. Glycopeptides resistivity is due to the alteration of targets.
- Both Gram-positive and Gram-negative organisms produce beta-lactamases. In Gram-positive organisms, penicillinase is the enzyme causing resistance towards antibiotics and can also cause a mutation by altering the enzymatic activity.
- Gram-negative organisms can express both chromosomal as well as plasmid-mediated enzymes that have broad activity in causing resistivity.
- Resistant to methicillin and oxacillin in S. aureus staphylococcal cassette chromosome mec which contains mec A gene. PBP2a is encoded by mec A gene which shows high resistance to beta-lactam antibiotics.
- Different antibiotics are exported from cells by membrane proteins to maintain their concentration called efflux pumps. These pumps carry a wide range of antibiotics that contribute to forming MDR organisms.
- D-alanyl-alanine is changed to D-alanyl-lactate which inhibits the cross-linking of glycopeptides hence causing resistance.
- Seven van genes are responsible for causing vancomycin resistance. These genes encode dehydrogenases that form lactate which is important for the formation of unmodified peptidoglycan.
- Characterization of different enzymes that are required for the transfer of plasmid results in vancomycin resistance.
References
- Canzani D and Aldeek, F. (2017). Penicillin G ‘s function, metabolites, allergy, and resistance. 1(1).
- Ghooi, R. B and Services SC. (2018). Inhibition of Cell Wall Synthesis -Is this the Mechanism of Action of Penicillins ? 9877(April), 2–7. https://doi.org/10.1016/0306-9877(95)90085-3
- Kang H and Park Y (2015). Glycopeptide Antibiotics : Structure and Mechanisms of Action. 45(2), 67–78.
- Kapoor G, Saigal S and Elongavan A (2017). Action and resistance mechanisms of antibiotics: A guide for clinicians. Journal of Anaesthesiology and clinical pharmacology. 33: 300-305
- Nikolaidis I and Dessen A (2014). Resistance to antibiotics targeted to the bacterial cell wall. 23, 243–259. https://doi.org/10.1002/pro.2414
- Scheffers, D and Pinho M G. (2005). Bacterial Cell Wall Synthesis : New Insights from Localization Studies. 69(4). 585–607. https://doi.org/10.1128/MMBR.69.4.585
- Singh M, Chang J, Coffman L, Kim S J and States U. (2018). HHS Public Access. 121(16), 3925–3932. https://doi.org/10.1021/acs.jpcb.7b00324.A
Sources
- https://www.researchgate.net/publication/268793620_Multidrug_efflux_pumps_in_Gramnegative_
bacteria_and_their_role_in_antibiotic_resistance- 6% - https://www.sciencedirect.com/topics/neuroscience/peptidoglycan- 4%
- https://www.sciencedirect.com/topics/immunology-and-microbiology/peptidoglycan- 3%
- https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/peptidoglycan- 2%
- https://www.sciencedirect.com/science/article/abs/pii/S0045206814000224- 1%
- https://globalrph.com/infectious-disease-list/monobactams/- 1%
- https://pubs.acs.org/doi/10.1021/acs.jpcb.7b00324- 1%
- https://pubs.asahq.org/anesthesiology/article/129/2/335/17980/Presumed-Lactam-Allergy-and-Crossreactivity-
in- 1% - https://docs.ufpr.br/~microgeral/Regulacaopeptideoglicana.pdf- 1%
