Interesting Science Videos
Modes of transmission of SARS-CoV-2 (COVID-19)
There are three modes of transmission of SARS-CoV-2 which include:
- Droplets transmission
- Contact transmission
- Aerosol transmission
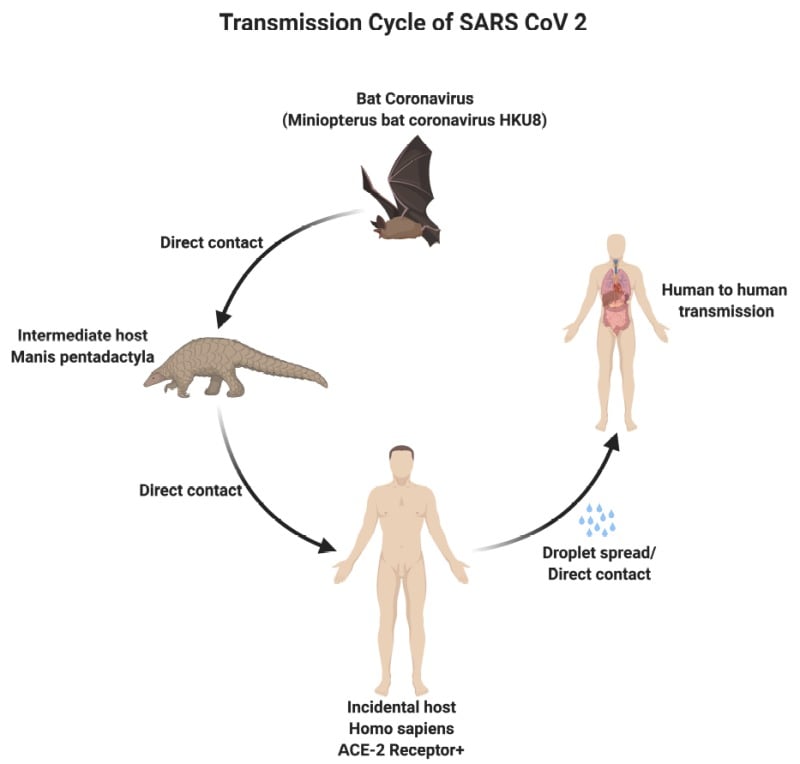
Figure: Modes of transmission of SARS-CoV-2. Image Source: NCBI Book
- Droplets transmission occurs when respiratory droplets (as produced when an infected person coughs or sneezes) are ingested or inhaled by individuals in close proximity.
- Contact transmission occurs when a subject touches a surface or object contaminated with the virus and subsequently touches their mouth, nose, or eyes.
- Aerosol transmission occurs when respiratory droplets mix into the air, forming aerosols, and cause infection while inhaling a high dose of aerosols into the lungs in a relatively closed environment.
Pathogenesis of SARS-CoV-2 (COVID-19)
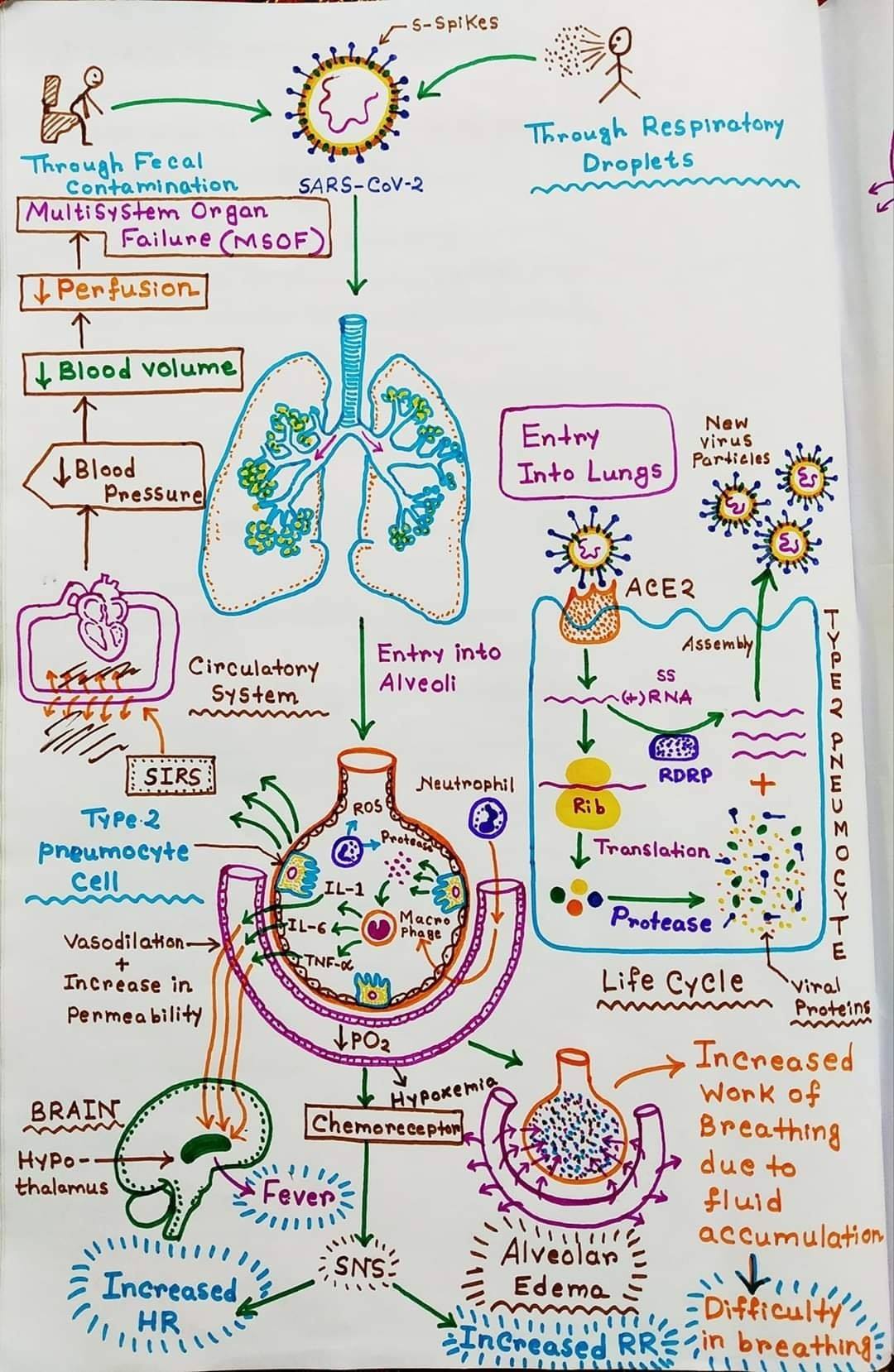
Figure: Mechanism of SARS-CoV-2 infection. Image Source: Soumik Roy. Concept By: Ninja Nerd Science.
- The incubation period ranges from 1 day to 14 days. The variation in incubation period is associated with the age of the patients above the age of 60 usually develop the symptoms faster as compared to others.
- SARS-CoV-2 affects the epithelial cells in the alveoli. After the entry of the virus to the respiratory tract, the cilia in the lower respiratory tract facilitate the attachment of the virus to its receptor.
- The cilia move the mucus covering the respiratory epithelium it moved upwards towards the throat. This facilitates the attachment of the virus.
- Multiple SARS-CoV-2 proteins interact with components of the innate immune system to evade an antiviral interferon response.
- The expression of hACE2, the SARS-CoV-2 receptor, on the surface of cells is downregulated after infection with SARS-CoV-2.
- The mechanism of this downregulation appears to be due to the internalization of ACE2 during SARS-CoV entry and by induction of TNFα converting enzyme activity of metalloproteases.
- The enzymes cleave the ACE2 extracellular domain from its transmembrane domain, resulting in shedding of this domain.
- This has led to the hypothesis that the binding of SARS-CoV-2 S protein is a virulence factor for SARS above and beyond its role in viral attachment and entry.
- Patients infected with COVID-19 show higher leukocyte numbers, abnormal respiratory findings, and increased levels of plasma pro-inflammatory cytokines.
- The main pathogenesis of COVID-19 infection as a respiratory system targeting virus is severe pneumonia, RNAaemia, combined with the incidence of acute cardiac injury.
- Significantly high blood levels of cytokines and chemokines are noted in patients with COVID-19 infection that include IL1-β, IL1RA, IL7, IL8, IL9, IL10, basic FGF2, GCSF, GMCSF, IFNγ, IP10, MCP1, MIP1α, MIP1β, PDGFB, TNFα, and VEGFA.
- Besides, high levels of pro-inflammatory cytokines including IL2, IL7, IL10, GCSF, IP10, MCP1, MIP1α, and TNFα that are reasoned to promote disease severity are observed in patients suffering from severe symptoms.
- The association of worsening clinical progression with declining virus loads and the onset of an immunological response, plus the presence of markedly elevated cytokines levels suggest that severe lung damage is largely immunopathological in nature.
COVID-19 Pathophysiology Video By Dr. Matt & Dr. Mike

Replication of Coronavirus (SARS-CoV-2)
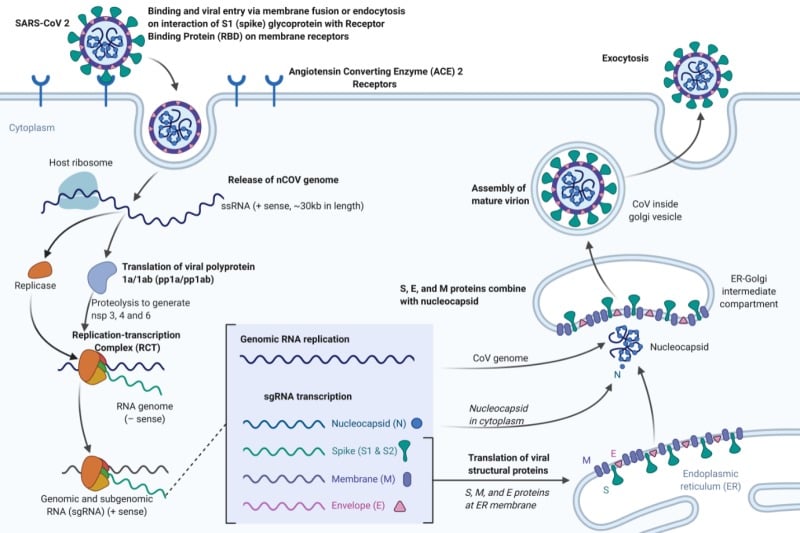
Figure: Replication of Coronavirus (SARS-CoV-2). Image Source: NCBI Book created with biorender.com
- It has been confirmed that the SARS-CoV-2 uses the same cellular entry receptor, ACE2, as SARS-CoV.
- Human ACE2, found in the lower respiratory tract of humans, works like the cell receptor for SARS-CoV and regulates both the cross-species and human-to-human transmission.
- The virion S-glycoprotein present on the surface of coronavirus attaches to the receptor, ACE2 on the surface of human cells.
- It has been shown that S protein and ACE2 binding efficiency is 10- to 20- fold higher than that of SARS-CoV.
- In the case of SARS-CoV, the cleavage of trimer S protein is initiated by the cell surface-associated transmembrane protease serine 2 (TMPRSS2) and cathepsin. At the same time, the possible molecules facilitated membrane invagination for SARS-CoV-2 endocytosis are still unclear.
- However, it has been observed that the SARS-CoV-2 may readily transmit while causing less severe human infection rather than human SARS-CoV.
- S glycoprotein includes two subunits, S1 and S2.
- S1 is responsible for the determination of the virus-host range and cellular tropism via Receptor Binding Domain (RBD), while S2 facilitates virus-cell membrane fusion by two tandem domains, heptad repeats 1 (HR1) and heptad repeats 2 (HR2).
- The genomic RNA of coronavirus of approximately 30,000 nucleotides encodes structural proteins and nonstructural proteins of the virus that have a critical role in viral RNA synthesis (called replicase-transcriptase proteins).
- At least one niche-specific protein, nonstructural protein 2 (nsp2), and one structural protein, the nucleocapsid protein (N), are involved in viral RNA synthesis.
- The expression of the coronavirus replicase-transcriptase protein genes is mediated by the translation of the genomic RNA.
- The replicase-transcriptase proteins are encoded in open-reading frame 1a (ORF1a) and ORF1b and are synthesized initially as two large polyproteins, pp1a and pp1ab.
- The synthesis of pp1ab involves programmed ribosomal frameshifting during translation of ORF1a.
- During or after synthesis, these polyproteins are cleaved by virus-encoded proteinases with papain-like (PLpro) and chymotrypsin-like folds into 16 proteins.
- NSP1 to NSP11 are encoded in ORF1a, and NSP12 to NSP16 are encoded in ORF1b.
- The replicase-transcriptase proteins, together with other viral proteins and, possibly, cellular proteins, assemble into membrane-bound replication-transcription complexes (RTC).
- These complexes accumulate at perinuclear regions and are associated with double-membrane vesicles.
- Hydrophobic transmembrane domains are present in NSP3, NSP4, and NSP6 likely serve to anchor the nascent pp1a/pp1ab polyproteins to membranes during the first step of RTC formation.
- Finally, the virion-containing vesicles fuse with the plasma membrane of the cell to release the virus.
- The virus then attaches a new cell, and the cycle is repeated.
References
- Caly L, Druce J, Catton M, Jans D and Wagstaff K. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research. 2020. 104787. ISSN 0166-3542, https://doi.org/10.1016/j.antiviral.2020.104787. (http://www.sciencedirect.com/science/article/pii/S0166354220302011)
- Kaye M. (2006). SARS-associated coronavirus replication in cell lines. Emerging infectious diseases, 12(1), 128–133. https://doi.org/10.3201/eid1201.050496
- Weiss, S. R., & Navas-Martin, S. (2005). Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiology and molecular biology reviews : MMBR, 69(4), 635–664. https://doi.org/10.1128/MMBR.69.4.635-664.2005
- Weiss SR and Leibowitz JL. Chapter 4 – Coronavirus Pathogenesis. Advances in Virus Research. Academic Press. Volume 81. 2011. Pages 85-164. ISSN 0065-3527. https://doi.org/10.1016/B978-0-12-385885-6.00009-2.
- Guo Y, Cae Q, Hong Z, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Millitary Medical Reasearch. (2020) 7:11. https://doi.org/10.1186/s40779-020-00240-0
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods in molecular biology (Clifton, N.J.), 1282, 1–23. https://doi.org/10.1007/978-1-4939-2438-7_1
- Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications (2020) 11:1620 | https://doi.org/10.1038/s41467-020-15562-9
- Cascella M, Rajnik M, Cuomo A, et al. Features, Evaluation and Treatment Coronavirus (COVID-19) [Updated 2020 Mar 20]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2020 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK554776/
Sources
- 11% – https://www.sinobiological.com/research/virus/coronavirus-replication
- 9% – https://www.sciencedirect.com/science/article/pii/S0896841120300469
- 6% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1797243/
- 5% – https://europepmc.org/articles/PMC1797243
- 4% – https://www.sciencedirect.com/science/article/pii/B9780123858856000092
- 4% – https://mmrjournal.biomedcentral.com/articles/10.1186/s40779-020-00240-0
- 3% – https://www.papausa.com/tretement-of-covid-19-novel-corona-virus/
- 3% – https://www.alexmanos.co.uk/covid-19/
- 2% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7068984/
- 2% – https://idpjournal.biomedcentral.com/track/pdf/10.1186/s40249-020-00646-x
- 2% – https://idpjournal.biomedcentral.com/articles/10.1186/s40249-020-00646-x
- 1% – https://www.pnas.org/content/early/2020/03/11/2002589117
- 1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6888126/
- 1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3700180/
- 1% – https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.0010039
- 1% – https://en.wikipedia.org/wiki/SARS-CoV-2
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3397359/
- <1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2292640/
- <1% – https://www.mysinustory.com/cilia.html
- <1% – https://www.cell.com/cell/fulltext/S0092-8674(20)30262-2
