- Coronaviruses (CoVs) are enveloped positive sense, single-stranded RNA viruses that belong to the subfamily Coronavirinae, family Coronavirdiae, order Nidovirales.
- Four genera of CoVs, namely, Alphacoronavirus (αCoV), Betacoronavirus (βCoV), Deltacoronavirus (δCoV), and Gammacoronavirus (γCoV), are distinguished.
- This group of viruses is of zoonotic origin with αCoV and βCoV found in bats and rodents while δCoV and γCoV are found in avian species.
Interesting Science Videos
Structure of SARS-CoV-2
- SARS CoV-2 virus is a betacoronavirus.
- They are enveloped, positive-sense, single-stranded RNA viruses of zoonotic origin.
- They are spherical to pleomorphic particles, measuring between 80 and 160 nm in length.
- SARS CoV-2 contains four structural proteins, namely envelope (E), spike (S), membrane (M), and nucleocapsid (N).
- The S, M, and E proteins together form the envelope of the virus. The M protein is the most abundant, mostly responsible for the shape of the envelope. The E protein is the smallest structural protein.
- The S and M proteins are also the transmembrane proteins that are involved in virus assembly during replication.
- N proteins remain associated with the RNA forming a nucleocapsid inside the envelope.
- Although N protein is largely involved in processes relating to the viral genome, it is also involved in other aspects of the CoV replication cycle (assembly and budding) and the host cellular response to viral infection.
- Polymers of S proteins remain embedded in the envelope giving it a crown-like appearance, thus the name coronavirus.
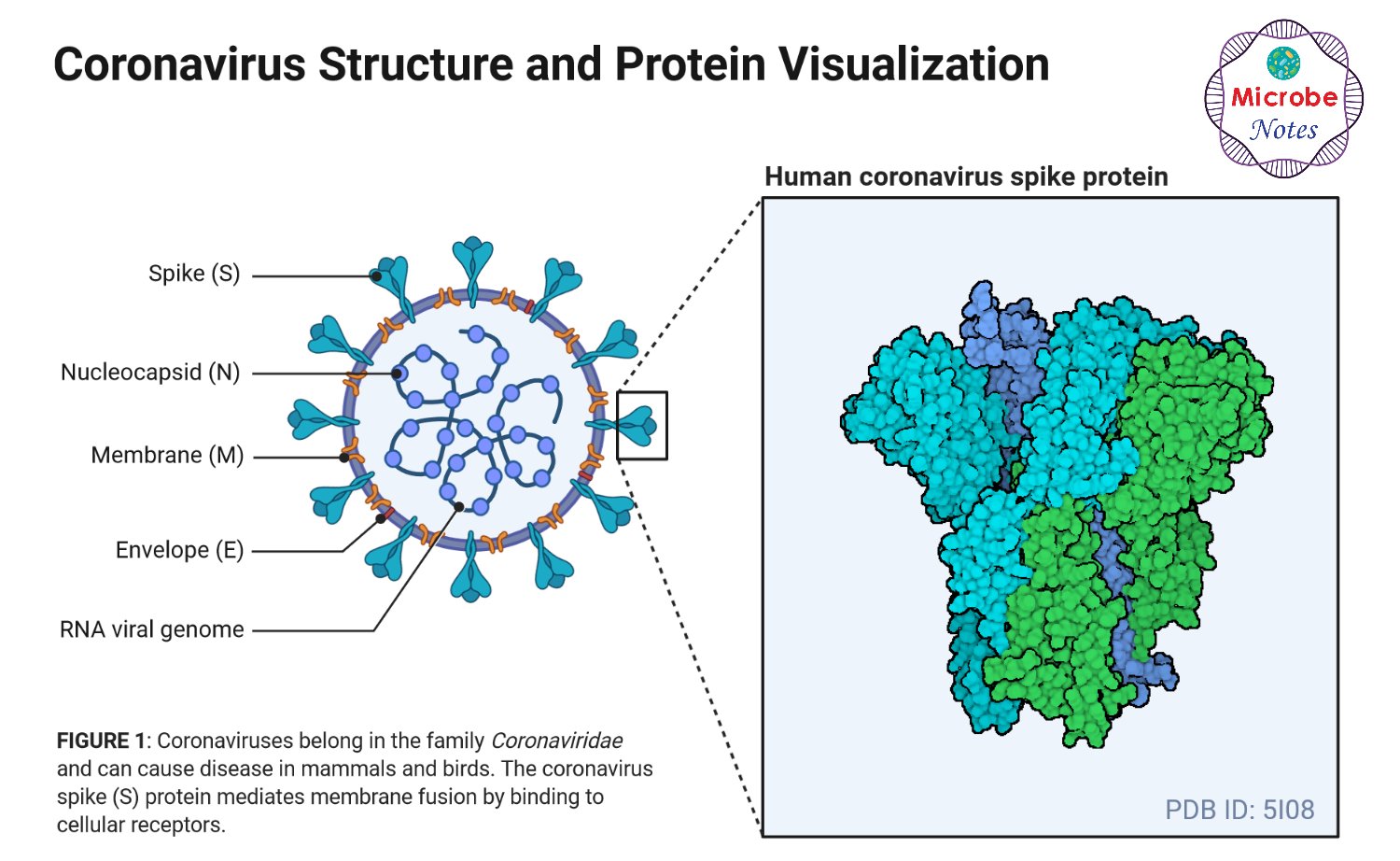
Figure: Structure of SARS-CoV-2, created with biorender.com
Spike Glycoprotein
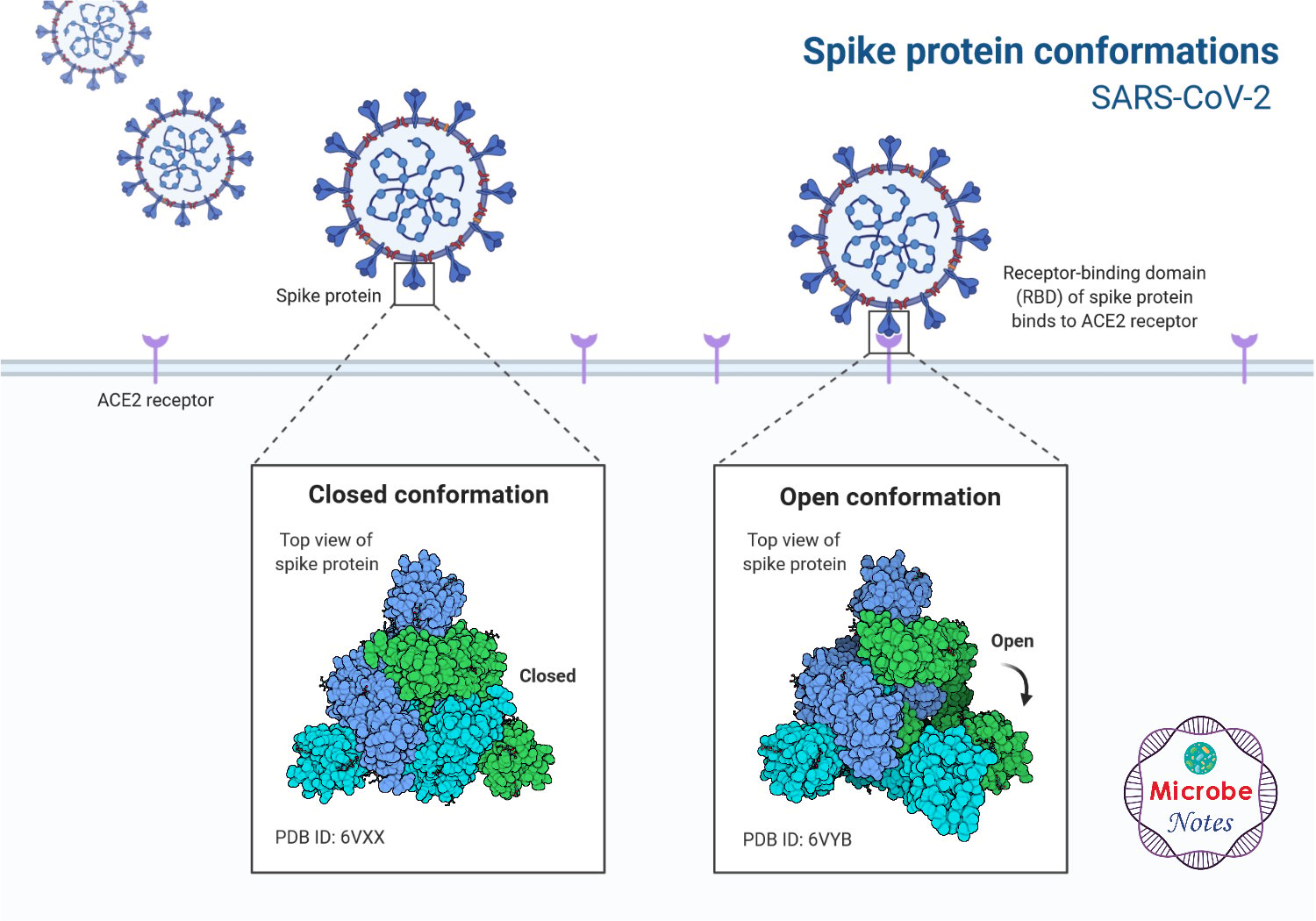
Figure: Spike protein conformation of SARS-CoV-2, created with biorender.com
- Spike glycoprotein comprised of S1 and S2 subunits. The S1 subunit contains a signal peptide, followed by an N-terminal domain and receptor-binding domain.
- The S2 subunit contains conserved fusion peptide, heptad repeat 1 and 2, a transmembrane domain, and a cytoplasmic domain.
- The S2 subunit of 2019-nCoV is highly conserved and shares 99% identity with those of the two bats SARS-like CoVs and human SARS-CoV.
- The S1 subunit shares 70% to these CoVs but the core receptor binding domain is highly conserved. These amino-acid differences are responsible for the direct interaction of spike protein with the host receptor.
- Spike glycoprotein binds to the human ACE2 receptor present in the target cells in the respiratory tract. This protein has a compact ridge that allows the virus to attach more strongly than other viruses of the same origin.
- After the spike protein binds with the receptor in the target cell, the viral envelope fuses with the cell membrane and releases the viral genome into the target cell.
Genomic Organization of SARS-CoV-2
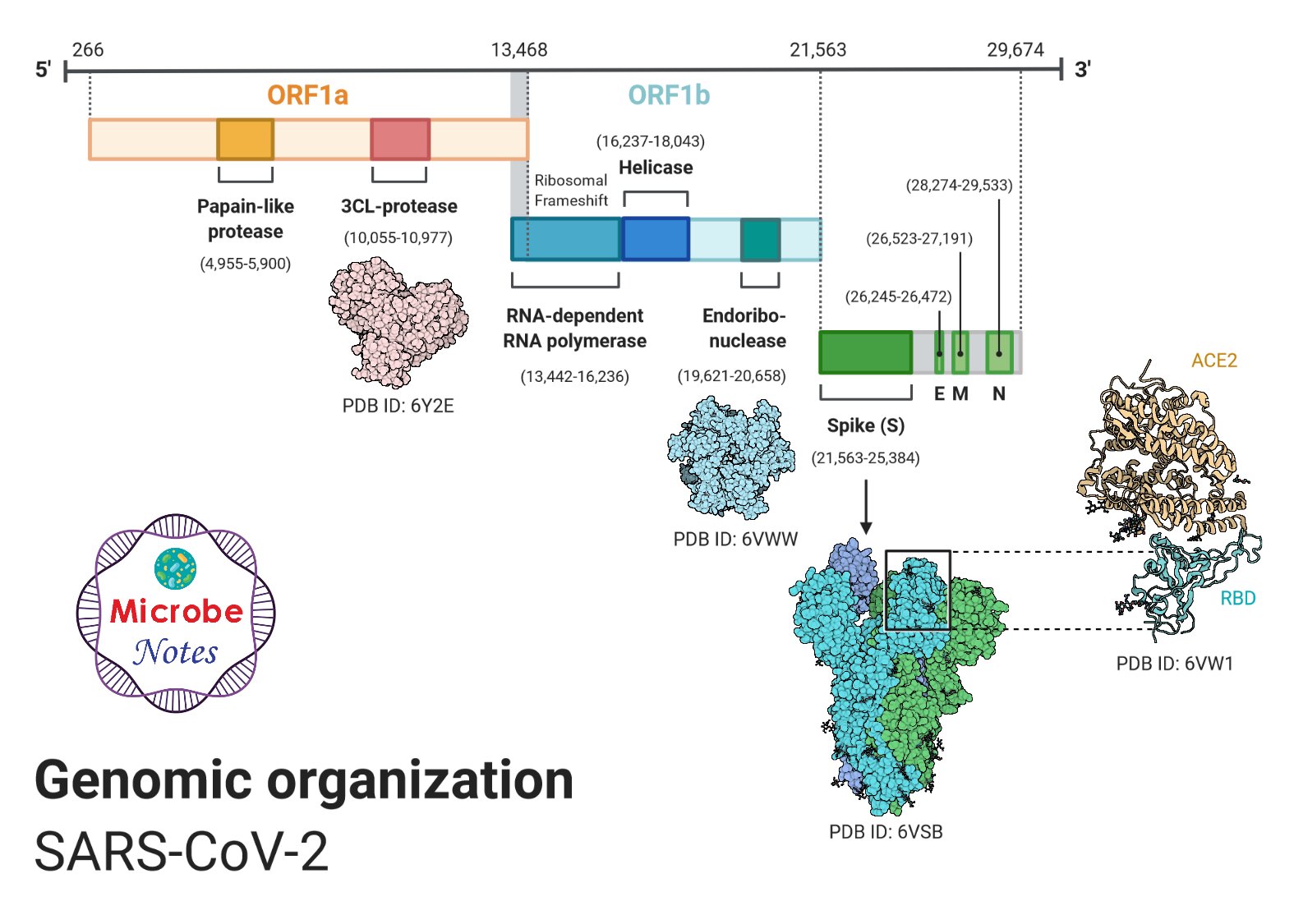
Figure: Genomic organization of SARS-CoV-2, created with biorender.com
- The genome of SARS-CoV-2 is a single-stranded positive-sense RNA of 30kb (29891 nucleotides) encoding 9860 amino acids. The G + C content is 38%.
- There are 12 functional open reading frames (ORFs) along with a set of nine subgenomic mRNAs carrying a conserved leader sequence, nine transcription-regulatory sequences, and 2 terminal untranslated regions.
- The genome of this virus lacks the haemagglutinin-esterase gene, which is characteristically found in lineage A βCoV.
- Two-thirds of viral RNA, mainly located in the first ORF translates two polyproteins, pp1a and pp1ab, and encodes 16 non-structural proteins (NSP), while the remaining ORFs encode accessory and structural proteins.
- The 16 non-structural proteins include two viral cysteine proteases, namely, NSP3 (papain-like protease) and NSP5 (main protease), NSP12 (RNA-dependent RNA polymerase, NSP13 (helicase), and other NSPs which are likely involved in the transcription and replication of the virus
- The rest part of the viral genome codes for four structural proteins E, M, S, and E along with a number of accessory proteins that interfere with the host immune response.
- The organization of the coronavirus genome is 5′-leader-UTR-replicase-S (Spike)–E (Envelope)-M (Membrane)-N (Nucleocapsid)-3′UTR-poly (A) tail with accessory genes interspersed within the structural genes at the 3′ end of the genome.
- SARS-CoV-2 is closer to the SARS-like bat CoVs in terms of the whole genome sequence.
- However, mutations are observed in NSP2 and NSP3 and the spike protein, that play a significant role in infectious capability and differentiation mechanism of SARS-CoV-2
- Besides, two strains, namely L-type and S-type, are discovered. It was found that L lineage was more prevalent than the S lineage within the limited patient samples that were examined. The study (https://academic.oup.com/nsr/article/7/6/1012/5775463) states that, “The implication of these evolutionary changes on disease etiology remains unclear”.
References
- Chan JF, Kok K, Zhu Z, Chu H, To KK, Yuan S and Yuen K (2020). Genomic characterization of the 2019 novel humanpathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan, Emerging Microbes & Infections, 9:1, 221-236, DOI: 10.1080/22221751.2020.1719902
- Xia S, Liu M, Wang C, et al.Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30, 343–355 (2020). https://doi.org/10.1038/s41422-020-0305-x
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods in molecular biology (Clifton, N.J.), 1282, 1–23. https://doi.org/10.1007/978-1-4939-2438-7_1
- Schoeman, D., Fielding, B.C. Coronavirus envelope protein: current knowledge. Virol J16, 69 (2019). https://doi.org/10.1186/s12985-019-1182-0
- Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications (2020) 11:1620 | https://doi.org/10.1038/s41467-020-15562-9
- Guo Y, Cae Q, Hong Z, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Millitary Medical Reasearch. (2020) 7:11. https://doi.org/10.1186/s40779-020-00240-0
- Adhikari et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infectious Diseases of Poverty. (2020) 9:29. https://doi.org/10.1186/s40249-020-00646-x
- https://www.livescience.com/why-coronavirus-attaches-stronger-human-cells.html
Sources
- 9% – https://mmrjournal.biomedcentral.com/articles/10.1186/s40779-020-00240-0
- 3% – https://www.tandfonline.com/doi/pdf/10.1080/22221751.2020.1719902
- 3% – https://www.researchgate.net/publication/264547924_The_Coronavirus_Nucleocapsid_Is_a_Multifunctional_Protein
- 3% – https://cmr.asm.org/content/cmr/28/2/465.full.pdf
- 2% – https://www.tandfonline.com/doi/full/10.1080/22221751.2020.1719902
- 2% – https://www.researchgate.net/publication/339176245_Fusion_mechanism_of_2019-nCoV_and_fusion_inhibitors_targeting_HR1_domain_in_spike_protein
- 1% – https://www.scientificamerican.com/article/researchers-map-structure-of-coronavirus-spike-protein/
- 1% – https://www.sciencedirect.com/science/article/pii/S0378111912011122
- 1% – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4369385/
- 1% – https://www.ncbi.nlm.nih.gov/books/NBK554776/
- 1% – https://www.cdc.gov/sars/lab/sequence.html
- 1% – https://taskboot.com/2020/03/05/new-coronavirus-mutants-misread-s-subtype-is-actually-the-ancestor-of-l-subtype/
- 1% – https://pubs.acs.org/doi/pdf/10.1021/acsinfecdis.0c00052
- 1% – https://bestpractice.bmj.com/topics/en-gb/3000168/aetiology
- 1% – https://atlanticbusinesssystems.com/2020/04/03/heres-why-the-new-coronavirus-is-so-good-at-infecting-human-cells/

Hola. Los dibujos son muy bonitos. O aporta foto al microscopio o no tienen credibilidad. Respecto al genoma de cerca de 30.000 nucleótidos ¿Cuántos pertenecen a algún material biológico y cuántos son producto de un constructo informático? En el nombre de la CIENCIA les exijo respuestas documentadas a éstas preguntas.
The final point needs to be updated.
‘Besides, two strains, namely L-type and S-type, are discovered. The L-type, derived from the S-type, is found to be more aggressive and contagious.’
The authors of the paper had to revoke this statement. The paper now states:
‘The implication of these evolutionary changes on disease etiology remains unclear.’
Thank you so much for the update. The information has been updated.
good work . all the things are undestood very easily.
Very good work on the structure of COVID -19 .Explaination is also simple and brief.I thank all who devoted for the work
a very good research work but please i still need the cumulative mechanism of action of the virus. thanks for the good work
Very nice explanation
Buena información de su origen y estructura molecular