Interesting Science Videos
What is Staphylococcus epidermidis?
Staphylococcus epidermidis is a Gram-positive bacterium belonging to the genus Staphylococcus and is the most frequently isolated species from human epithelia.
- Staphylococci are known as clustering Gram-positive cocci, nonmotile, non-spore-forming facultatively anaerobic that classified into two main groups, coagulase-positive and coagulase-negative.
- S. epidermidis belongs to the group of coagulase-negative staphylococci (CoNS), which is different from coagulase-positive staphylococci such as S. aureus by lacking the enzyme coagulase.
- It colonizes the axillae, head, and nares predominantly and is assumed to provide protection from the harsh conditions that might be present in those habitats.
- This bacterium was distinguished from S. aureus in 1884 by Friedrich Julius Rosenbach, initially naming it S. albus. It was later renamed by Evans in 1916.
- S. epidermidis is mostly non-pathogenic but might cause infections in patients with a compromised immune system. Thus, most of these infections are hospital-acquired.
- It is a part of the human normal flora as it resides over the epidermis of the skin.
Classification of Staphylococcus epidermidis
Classification of species of the genus Staphylococcus is based on various factors like the chemical properties of the cell wall, especially the amino acid composition and sequence of the interpeptide bridges of the peptidoglycan and teichoic acid composition. Besides, other biochemical properties of staphylococci are useful for the differentiation of species, including the production of lactic acid when grown under anaerobic conditions.
The following is the taxonomical classification of S. epidermidis based on various morphological, physiological, and biochemical characteristics;
Domain: Bacteria
Phylum: Firmicutes
Class: Bacilli
Order: Bacillales
Family: Staphylococcaceae
Genus: Staphylococcus
Species: S. epidermidis
Habitat of Staphylococcus epidermidis
- S. epidermidis is the predominant coagulase-negative Staphylococcus species found in the material of human origin. Humans are the only natural host for this organism.
- The physiological habitat of S. epidermidis is the skin and mucous membranes of humans and animals. The name ‘epidermidis’ indicates the habitat of the organism.
- S. epidermidis is the most familiar resident staphylococcal species on human skin in terms of population size.
- It is a ubiquitous inhabitant of human skin and mucous membranes that forms a part of the normal flora of skin in humans, predominantly found in the nasal passage and sweaty areas of the body like the armpits and the back.
- It is also the primary resident of the scalp, different areas throughout the face, groin, and legs.
- The largest populations of cutaneous staphylococci (104–106 CFU/cm2) are found in regions of the skin of mammals supplied with large numbers of pilosebaceous units and sweat glands and on the skin and mucous membranes surrounding openings to the body surface.
- The exact physiological role of the bacterium as the normal flora is not yet known; however, it is assumed that it may be involved in the lipid metabolism of the skin and that it may serve as a primary barrier against invading microbial pathogens.
- Besides, these are also considered responsible for body odor caused due to various secretions.
Morphology of Staphylococcus epidermidis
- S. epidermidis is a Gram-positive bacterium that appears spherical with an average diameter of 0.5–1.5 µm on light microscopy. The cells of old cultures (>48 h) are often gram-variable to nearly gram-negative.
- Cells divide in more than one plane to form irregular clusters and aggregates of pairs, tetrads, and short chains.
- Cells of S. epidermis are considered cell wall deficient or defective (L-form), which is why these organisms fail to take Gram strain, are osmotically sensitive, and are not easily cultured on the usual isolation media.
- The cell membrane is a typical lipid-protein bilayer, composed mainly of phospholipids and proteins. Phospholipids, glycolipids, menaquinones, and carotenoids make up the major lipid components of the membrane.
- Some members of the S. epidermidis group are known to express certain iron-regulated cell membrane proteins are expressed under iron limitation.
- Besides, a 42 kDa cell wall protein that binds human transferrin, the major iron-binding protein in serum, has been detected in some members of this species.
- The cell wall is made up of peptidoglycan and teichoic acid, where the peptidoglycan is the main structural polymer in the wall and plays a vital role in maintaining the spherical shape of the cell.
- Few wall-associated proteins have also been seen in S. epidermidis, some of which are fibronectin-binding proteins.
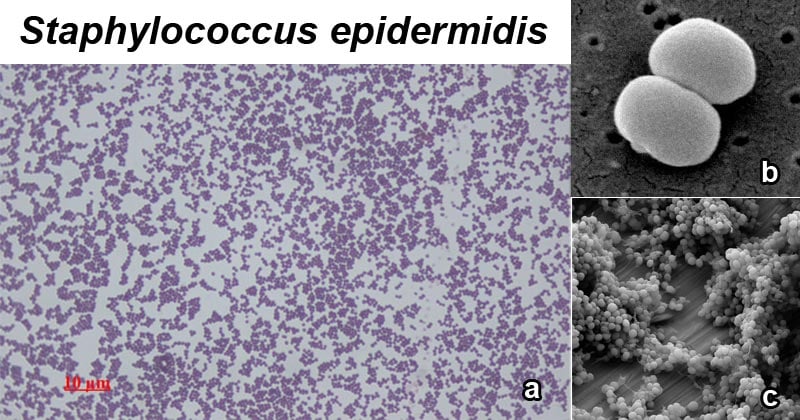
Figure: a. Staphylococcus epidermidis, 1000 magnification under bright field microscopy; b. Scanning electron image of S. epidermidis; c. Staphylococcus epidermidis biofilm on titanium substrate. Image Source: Wikipedia.
Cultural characteristics of Staphylococcus epidermidis
- Staphylococci produce distinctive colonies on a variety of commercial, selective, and nonselective agar media.
- The commonly used selective media include mannitol–salt agar, lipase–salt–mannitol agar, phenyl ethyl alcohol agar, Columbia colistin–nalidixic acid (CNA) agar, and Baird–Parker agar base supplemented with egg yolk tellurite enrichment.
- Colonies of the same strain generally exhibit similar features of size, consistency, edge, profile, luster, and pigment, but some strains may produce two or more morphotypes.
- Especially with S. epidermidis, the variable expression of the gene coding for PIA is considered to cause the variation in morphologies among the same strain.
- The following are the colony morphologies of S. epidermidis on different agar media:
1. Nutrient Agar (NA)
- Circular, cream-colored to white colonies of S. epidermidis are observed on NA. The colonies are mostly 1mm in diameter with an entire margin.
- The colonies have raised elevation and a dense center with transparent borders.
2. Mannitol Salt Agar (MSA)
- Small pink to red colonies are formed on MSA. The media remains red as the bacterium cannot ferment mannitol.
- This media is a selective media for S. aureus and is commonly used to distinguish S. aureus from S. epidermidis.
3. Tryptic soy agar (TSA)
- On tryptic soy agar, S. epidermidis produces white raised, cohesive colonies of the size 1-2 mm in diameter.
- Some strains of S. epidermidis are also known to produce subtle violet, pinkish, or brownish pigment.
4. Congo Red Agar (CRA)
- Pale pink to red and opaque colonies with a rough texture and a central nipple is seen on CRA.
Biochemical Characteristics of Staphylococcus epidermidis
The biochemical characteristics of S. epidermidis can be tabulated as follows:
| S.N | Biochemical Characteristics | S. epidermidis |
| 1. | Capsule | Most of the strains are capsulated. |
| 2. | Shape | Cocci |
| 3. | Catalase | Positive (+) |
| 4. | Oxidase | Negative (-) |
| 5. | Citrate | Negative (-) |
| 6. | Methyl Red (MR) | Negative (-) |
| 7. | Voges Proskauer (VR) | Negative (-) |
| 8. | Urease | Positive (+) |
| 9. | Coagulase | Negative (-) |
| 10. | Gas | Positive (+) |
| 11. | H2S | Positive (+) |
| 12. | Hemolysis | Negative (-) |
| 13. | Motility | Negative (-) |
| 14. | Nitrate Reduction | Positive (+) |
| 15. | Gelatin Hydrolysis | Negative (-) |
| 16. | Pigment Production | Negative (-) |
Fermentation Test
| S.N | Substrate | S. epidermidis |
| 1. | Mannitol | Positive (+) |
| 2. | Glucose | Positive (+) Lactate is the major end product of anaerobic glucose metabolism. |
| 3. | Fructose | Positive (+) |
| 4. | Galactose | Variable |
| 5. | Lactose | Positive (+) |
| 6. | Maltose | Positive (+) |
| 7. | Mannose | Positive (+) |
| 8. | Raffinose | Negative (-) |
| 9. | Ribose | Variable |
| 10. | Sucrose | Positive (+) |
| 11. | Starch | Negative (-) |
| 12. | Trehalose | Negative (-) |
| 13. | Xylose | Negative (-) |
| 14. | Salicin | Negative (-) |
| 15. | Glycerol | Negative (-) |
| 16. | DNase | Negative (-) |
| 17. | Dulcitol | Negative (-) |
| 18. | Cellobiose | Negative (-) |
| 19. | Rhamnose | Negative (-) |
| 20. | Arabinose | Negative (-) |
| 21. | Inulin | Negative (-) |
Enzymatic Reactions
| S.N | Enzymes | S. epidermidis |
| 1. | Hyaluronidase | Variable |
| 2. | Acetoin | Positive (+) |
| 3. | Alkaline Phosphatase | Positive (+) |
| 4. | Ornithine Decarboxylase | Variable |
| 5. | Tryptophan Deaminase | Negative (-) |
| 6. | Arginine Dehydrolase | Negative (-) |
- Besides, Members of the S. epidermidis species group have numerous amino acid requirements. They require arginine, isoleucine–valine, and proline.
- Vitamin requirements have also been determined in some members of S. epidermidis as they require nicotinic acid, thiamine, biotin, and pantothenic acid.
Virulence factors of Staphylococcus epidermidis
Even though S. epidermidis has not evolved to cause diseases, it is now frequently associated with various nosocomial infections. The organism is also provided with several mechanisms to evade being ingested and killed by the host immune system. Some of these mechanisms and products that protect the organism from the host immune defenses during infections are:
1. Biofilm formation
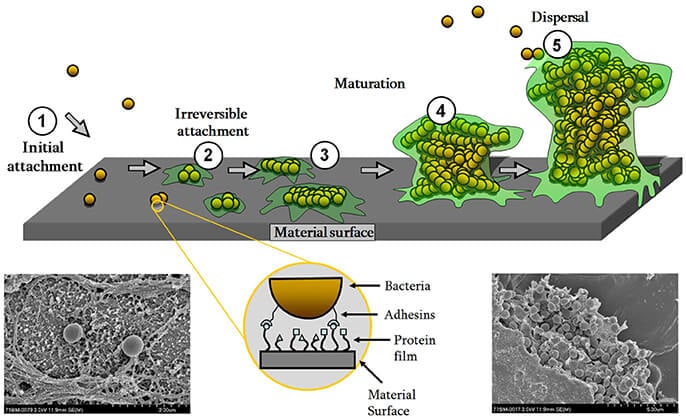
Figure: Biofilm formation scheme with scanning electron micrographs of S. epidermidis single cells (lower left) or in the biofilm community surrounded by EPS (lower right) on a titanium surface. Image Source: https://doi.org/10.3389/fmicb.2017.01401.
- Biofilm formation is the most important virulence factor of S. epidermidis to cause various acquired infections. It supports colonization, immune evasion as well as antibiotic resistance of the organism.
- Biofilms are multicellular aggregates of microorganisms enclosed in the extracellular matrix either produced by the organism or derived from the environment.
- The formation of a biofilm provides an advantage to the organism where it can better adapt to the environmental factors and has increases resistance to hostile conditions.
- Biofilm formation is achieved via two steps, the first being initial attachment of the bacteria followed by the subsequent aggregation and maturation into multicellular structures.
- S. epidermidis has surface proteins like AtlE, which is a bifunctional adhesin/autolysin, and the Bap/Bhp protein which contributes to the adhesion of bacteria to biotic and abiotic surfaces.
- Besides, S. epidermidis also has various surface proteins called MSCRAMMs (Microbial surface components recognizing adhesive matrix molecules) with the ability to interact with matrix proteins and support colonization.
- The most important MSCRAMM of S. epidermidis is SdrG (Fbe), a fibrinogen-binding protein that is necessary and sufficient to promote S. epidermidis adhesion to fibrinogen on the skin surfaces.
- After initial adhesion, biofilms develop via intercellular aggregation that is mediated by many different surface macromolecules like a poly-N-acetylglucosamine (PNAG) homopolymer, also named PIA (polysaccharide intercellular adhesin).
2. Protective exopolymers
- S. epidermidis produces exopolymers, namely poly-γ-glutamic acid (PGA) and PNAG/PIA that protect from important mechanisms of innate host defense.
- This exopolysaccharide has also been found to protect S. epidermidis from neutrophil killing, complement deposition, and immunoglobulins.
3. Pathogen-associated molecular patterns
- Additionally, several additional molecules that are specific to S. epidermidis are also found to stimulate innate host defense and thus act as pathogen-associated molecular patterns (PAMPs).
- The PNAG/PIA is reported to stimulate the Toll-like receptor 2 which represents an interesting example of the hide-and-seek interplay between pathogen and host, as a substance that S. epidermidis uses for immune evasion would trigger innate host defense mechanisms.
Pathogenesis of Staphylococcus epidermidis
Infections caused by S. epidermidis have been found to cause invasive infections in selected groups of patients, which include preterm neonates, immunocompromised individuals, and patients with permanent medical devices. The pathogenesis of infection caused by S. epidermidis can be explained as follows:
1. Adhesion/ Attachment/ Colonization
- As a commensal microorganism, S. epidermidis retains the ability to adhere to host proteins in the skin specifically.
- In a surgical wound, the bacterium utilizes these adhesion mechanisms to adhere to the deeper tissues and to the implanted device.
- Initial adhesion of bacteria to implant surfaces is mediated by non-specific interactions by specific adhesins such as autolysin extracellular DNA.
- In the context of medical devices, the surface of the device becomes coated with host-derived plasma proteins, extracellular matrix proteins, and coagulation products (platelets and thrombin) immediately after the following adhesion.
- The initial adhesion is then followed by irreversible attachment which is brought about by SdrG (Fbe), a fibrinogen-binding protein that promotes the binding of S. epidermidis to fibrinogen on the skin surfaces.
- In the case of medical devices, wall teichoic acid enhances the initial adhesion of S. epidermidis to medical devices by binding to adsorbed fibronectin.
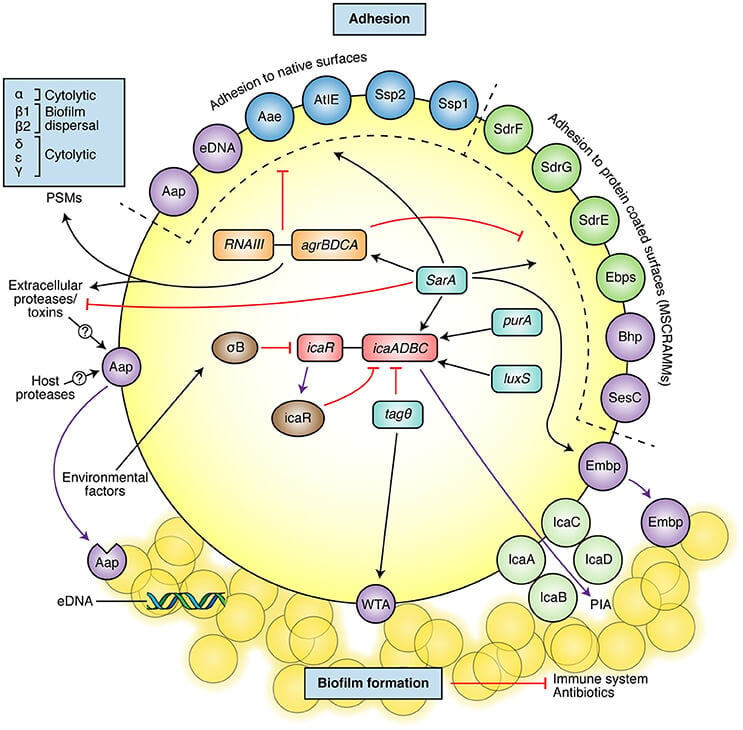
Figure: Scheme of the main S. epidermidis pathogenic mechanisms, which include adhesion molecules and biofilm formation. The most well-described adhesins involved in adhesion to native surfaces or protein-coated surfaces are shown in the upper part (molecules also involved in biofilm formation shown in purple). The main described biofilm components are shown at the bottom of the figure (PIA, cleaved Aap, eDNA, WTA, and Empb). The figure also presents some of the most important regulators of biofilm and adhesion molecules (black arrows: activation/positive signaling, red lines: inhibition/negative signaling). Image Source: https://doi.org/10.3389/fmicb.2017.01401.
2. Biofilm Formation
- The initial adhesion of bacteria to the biotic and abiotic surfaces is the first step of biofilm formation, the most important virulence factor of the organism.
- The accumulation and maturation of the S. epidermidis biofilm occur via a number of mechanisms.
- Polysaccharide intercellular adhesin (PIA, or poly-N-acetyl-glucosamine (PNAG)), is responsible for biofilm formation in the majority of S. epidermidis isolates.
- In others, however, biofilm formation is brought about by proteinaceous factors, such as the accumulation associated protein (Aap).
- Biofilms play a role in immune evasion, primarily by providing a barrier to immune cells.
- Further, PIA contributes to innate immune system evasion by promoting the generation of complement C5a fragment, inhibiting phagocytes and neutrophil killing, and reducing the activity of AMPs.
- The bacterial cells within the biofilm are embedded in an exopolysaccharide matrix which allows the bacterial population protection from host defense mechanisms and antimicrobial agents.
3. Dispersal
- Once the biofilms are formed, the bacteria present in the biofilms tend to disperse out to new secondary infections sites.
- The characteristic structure of mature biofilms with mushroomlike shapes and channels is dependent on the production of cell-cell disruptive factors, which in S. epidermidis are phenol-soluble modulins (PSMs).
- These surfactant-like molecules presumably work by decreasing non-covalent adhesion between cells, which causes dissemination of biofilm-associated infection, cells, or cell clusters may detach from a biofilm to reach secondary infection sites.
- The clinical symptoms that result from biofilm-related infections are mostly caused due to the host immune response to antigens released from the biofilm.
- However, the host immune response might cause more damage to the surrounding tissues instead of clearing the biofilm.
Clinical Manifestations of Staphylococcus epidermidis
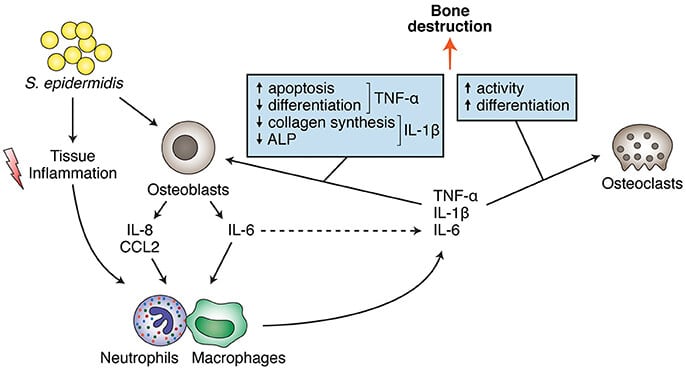
Figure: S. epidermidis direct and indirect effects on bone cells (osteoblasts and osteoclasts), leading to bone destruction. Image Source: https://doi.org/10.3389/fmicb.2017.01401.
- S. epidermidis is primarily associated with nosocomial infections which are primary associated with permanent medical implant devices like catheters.
- This bacterium is associated with more than 22% of the bloodstream infections of the central intravenous catheters.
- Besides, this microorganism may play a significant role in shunt, prosthetic joint, vascular graft, and surgical site infections.
- Eye keratitis and endophthalmitis of contaminated contact lens, urinary catheter infections, bacteremia, mediastinitis, and other infections are associated with S. epidermidis.
- S. epidermidis infections are seldom lethal, but they significantly contribute to morbidity and health care costs.
- Other infections include pneumonia, deep abscesses, osteomyelitis, endocarditis, phlebitis, mastitis, and meningitis, and are often associated with hospitalized patients rather than healthy individuals in the community.
Lab Diagnosis of Staphylococcus epidermidis
As with most bacterial infections, the collection of clinical specimens is the first step of laboratory diagnosis. In the case of S. epidermidis, clinical specimens like the scabs, joint aspirates, and pus aspirated from deep sites are to be collected. Diagnosis of disease in the case of S. epidermidis infections are mostly related to the identification of the organism. Some of the methods of identification are given below:
1. Microscopic observation and Biochemical tests
- Direct microscopic examination of these specimens may provide a rapid, presumptive report of gram-positive cocci resembling staphylococci.
- Direct observation is followed by isolation of the organism from primary clinical specimens on selective culture media like tryptic soy agar supplemented with 5 percent sheep blood), following an incubation period of 18–24 h in the air at 35–37°C.
- Initial identification can be made by observing the colonies on the culture media. The isolated colonies can then be subjected to various biochemical tests.
- Depending on the microscopic observation, colony morphology, and biochemical tests, S. epidermidis can be detected.
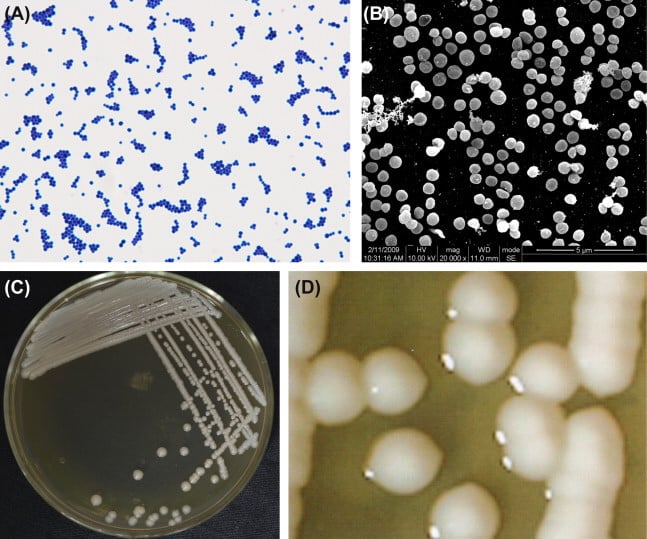
Figure: (A) Staphylococcus epidermidis cells (Gram stain). (B) S. epidermidis cells (SEM). S. epidermidis cells are spherical (0.5–1.5 μm in diameter) and gram-positive. The cocci organize into tetrads and clusters. Single cells are occasionally observed. (C) Colonies of S. epidermidis incubated on an agar plate. (D) Colonies of S. epidermidis (stereomicroscope). Image Source: Atlas of Oral Microbiology.
2. Rapid Identification kits
- Several rapid identification kits and automated systems can be found that are capable of identification of most species and subspecies within a few hours to one day with an accuracy of 70–90 percent.
- These kits are based on various factors like antibiotic susceptibility, enzyme production, immunological reactions, and cellular fatty acid analysis.
3. Molecular methods
- Molecular methods usually include tests that help in the identification of the organism at a molecular level.
- One of the most important molecular methods is Polymerase Chain Reaction (PCR) which helps in the amplification and detection of bacterial DNA.
- Besides, DNA sequencing can also be done to determine the DNA sequence of the bacteria that can then be used for its identification.
- Another essential and successful diagnostic method is the analysis of ribosomal RNA by the application of restriction fragment length polymorphisms (RFLP) methods.
Treatment of Staphylococcus epidermidis
- Because most S. epidermidis infections are associated with medical implant devices, the removal of these devices is the first mode of treatment.
- Other in-vitro infections are treated by the administration of antibiotic drugs that include cephalosporins like cefazolin, nafcillin, or oxacillin.
- For severe infections or antibiotic-resistant strains, Vancomycin is the drug of choice against these infections.
- Despite antibiotic treatment and elimination of related infection factors, medical implant infections are extremely resistant to antimicrobial agents.
- New approaches are being adopted to find a new set of antimicrobial targeting different molecules. Potential targets such as enzymes involved in an essential function are identified, and screening methods are then developed to identify inhibitors of the specific target molecule.
- Other forms of treatments involving the hyperimmune serum from human donors or humanized monoclonal antibodies directed towards the surface components are also being studied.
Prevention of Staphylococcus epidermidis
The inherent resistance of bacterial biofilms to antimicrobial agents, together with the increasing number of antibiotic-resistant strains, represents the need for effective preventive strategies. The following are some preventive strategies that can be followed to avoid such infections:
- Coating of biomaterials or use in exit-site dressings can be employed to prevent medical devices related to infections.
- Regular cleaning and dressing wounds might also work to prevent Staph infections to a certain extent.
- The use of aseptic techniques to prevent bacterial contamination from the insertion site and catheter hubs during insertions can also be applied.
Staphylococcus epidermidis video lecture by Dirty Medicine.
References
- Topley WWC (2007). Topley and Wison’s Microbiology and Microbial Interactions; Bacteriology, 2 Vol. Tenth Edition. John Wiley and Sons Ltd.
- Hildegunn Norbakken Granslo, Claus Klingenberg, Elizabeth Gladys Aarag Fredheim, Arild Rønnestad, Tom Eirik Mollnes, Trond Flægstad. The Arginine Catabolic Mobile Element is associated with low antibiotic resistance and low pathogenicity in Staphylococcus epidermidis from neonates. Pediatric Research 2010; 68: 237-41
- Elizabeth G. Aarag Fredheim, Hildegunn Norbakken Granslo, Trond Flægstad, Yngve Figenschau, Holger Rohde, Irina Sadovskaya, Tom Eirik Mollnes, Claus Klingenberg. Staphylococcus epidermidis Polysaccharide Intercellular Adhesin Activates Complement FEMS Immunology and Microbiology 2011; 63: 269–280
- Hildegunn Norbakken Granslo, Claus Klingenberg, Elizabeth Aarag Fredheim, Ganesh Acharya, Tom Eirik Mollnes, Trond Flægstad. Staphylococcus epidermidis biofilms induce lower complement activation in neonates compared to adults. Submitted to Infection and Immunity October 27th 2011.
- Peters G., Schumacher-Perdreau F., Jansen B. (1990) Staphylococcus epidermidis— a Versatile Pathogen. Pathogenesis of Wound and Biomaterial-Associated Infections. Springer, London. https://doi.org/10.1007/978-1-4471-3454-1_37
- Otto, M. Staphylococcus epidermidis— the ‘accidental’ pathogen. Nat Rev Microbiol 7, 555–567 (2009). https://doi.org/10.1038/nrmicro2182
- Namvar A E et al (2014). Clinical characteristics of Staphylococcus epidermidis: a systematic review. GMS Hyg Infect Control. 2014; 9(3): Doc23.
- Deighton, M., Capstick, J., & Borland, R. (1992). A study of phenotypic variation of Staphylococcus epidermidis using Congo red agar. Epidemiology and Infection,109(3), 423-432. doi:10.1017/S095026880005041X
- BRUCE E. LANGLOIS, ABDUL KARIM PARLINDUNGAN, ROBERT J. HARMON, KATHERINE AKERS; Biochemical Characteristics of StaphylococcusSpecies of Human and Bovine Origin. J Food Prot 1 February 1990; 53 (2): 119–126. doi: https://doi.org/10.4315/0362-028X-53.2.119
- Sabaté Brescó Marina, Harris Llinos G., Thompson Keith, Stanic Barbara, Morgenstern Mario, O’Mahony Liam, Richards R. Geoff, Moriarty T. Fintan. Pathogenic Mechanisms and Host Interactions in Staphylococcus epidermidis Device-Related Infection. Frontiers in Microbiology. 8. 2017. Pg: 1401. 10.3389/fmicb.2017.01401
- Paulus H. S.Kwakman, Anje A. te Velde, Christina M. J. E. Vandenbroucke-Grauls, Sander J. H. van Deventer, Sebastian A. J. Treatment and Prevention of Staphylococcus epidermidis Experimental Biomaterial-Associated Infection by Bactericidal Peptide 2. Antimicrobial Agents and Chemotherapy Nov 2006, 50 (12) 3977-3983; DOI: 10.1128/AAC.00575-06
- Foster T. Staphylococcus. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 12.Available from: https://www.ncbi.nlm.nih.gov/books/NBK8448/
- Doškař J., Pantůček R., Růžičková V., Sedláček I. (2010) Molecular Diagnostics of Staphylococcus aureus. In: Viola Magni M. (eds) Detection of Bacteria, Viruses, Parasites and Fungi. NATO Science for Peace and Security Series A: Chemistry and Biology. Springer, Dordrecht. https://doi.org/10.1007/978-90-481-8544-3_7
- Gaszewska-Mastalarz A, Mordarski M, Zakrzewska-Czerwińska J. Molekularna diagnostyka mikroorganizmów–identyfikacja Staphylococcus epidermidis [Diagnostic molecular microbiology–identification of Staphylococcus epidermidis]. Postepy Hig Med Dosw. 1998;52(1):19-34.
- O’gara JP and Humphreys H (2001). Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. Vol. 50 (2001). 582-587.
- http://www.bacteriainphotos.com/staphylococcus_epidermidis_colony_morphology.html

