Interesting Science Videos
Structure of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)
- They are enveloped viruses with round and sometimes pleiomorphic virions of approximately 80 to 120 nm in diameter.
- It contains positive-strand RNA, with the largest RNA genome (approximately 30 kb).
- The genome RNA is complexed with the basic nucleocapsid (N) protein to form a helical capsid found within the viral membrane.
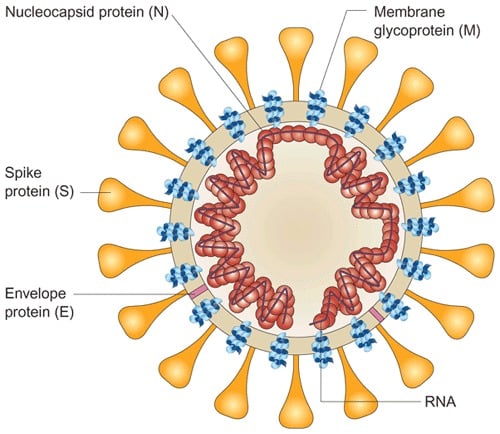
- The membranes of virus contains at least three viral proteins.
- These are spike (S), the type I glycoprotein that forms the peplomers on the virion surface, giving the virus its corona- or crown-like morphology in the electron microscope.
- The membrane (M) protein, a protein that spans the membrane three times and has a short N-terminal ectodomain and a cytoplasmic tail; and small envelope membrane protein (E), a highly hydrophobic protein.
- Hemagglutinin esterase (HE) is not encoded in the SARS-CoV genome.
Genome of SARS-CoV
- SARS Coronavirus genomes are monopartite, single-stranded, positive-sense, polyadenylated, and capped RNAs ranging from 27 to 32 kb in length.
- The 5′ approximately 20 to 22 kb carries the replicase gene, which encodes multiple enzymatic activities.
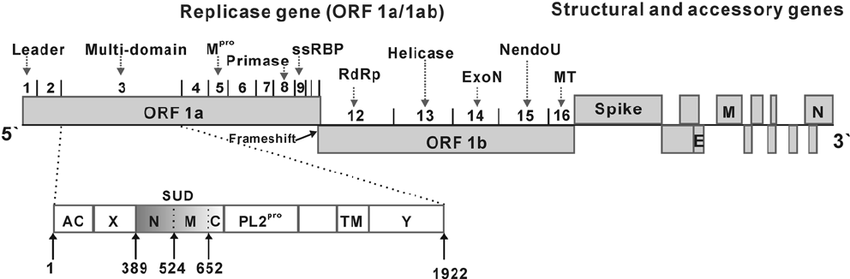
- The replicase gene products are encoded within two very large open reading frames, ORFs 1a and 1b, which are translated into two large polypeptides, pp1a and pp1ab, via a frameshifting mechanism.
- The structural proteins are encoded within the 3′ one-third of the genome, in the order S-E-M-N.
- There are untranslated regions (UTRs) on both the 5′ and 3′ ends of the genome, which are believed to interact with host and perhaps viral proteins to control RNA replication, which includes the synthesis of positive- and negative-strand genomic-length RNA.
Epidemiology of SARS-CoV
- SARS-CoV emerged in the human population in Guangdong province of southeastern, China in 2002, causing a worldwide epidemic with severe morbidity and high mortality rates with dramatic economic and societal consequences, particularly in older individuals.
- Within months, the outbreak had spread to 29 countries and regions causing over 8000 human cases and almost 800 deaths in the 2002-2003 outbreak.
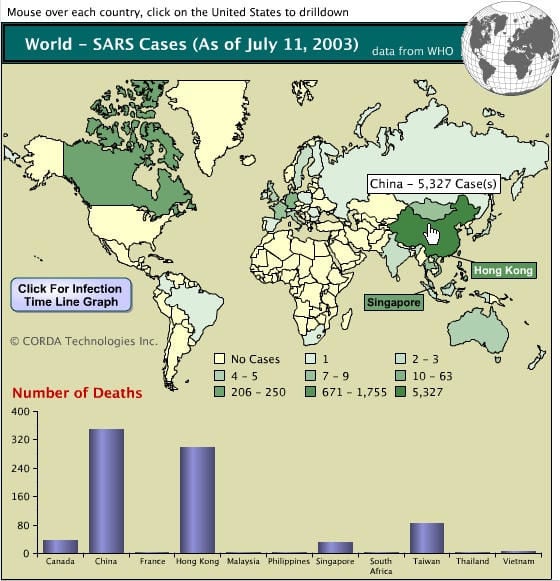
- SARS CoV emerged from a precursor virus which is endemic in insectivorous bats.
- The close proximity of different animal species (including bats) within large live game animal markets which service the restaurant trade for exotic food in southern China allowed the bat SARS CoV-like precursor virus to adapt to other mammalian species (civet cats, raccoon dogs) and subsequently, to humans.
Replication of SARS-CoV
- The replication cycle takes place in the cytoplasm of cells.
- The virus attaches to receptors on target cells by the glycoprotein spikes on the viral envelope.
- The functional receptor for SARS-CoV is angiotensin converting enzyme 2.
- The particle is then internalized, probably by absorptive endocytosis.
- The S glycoprotein may cause fusion of the viral envelope with the cell membrane.
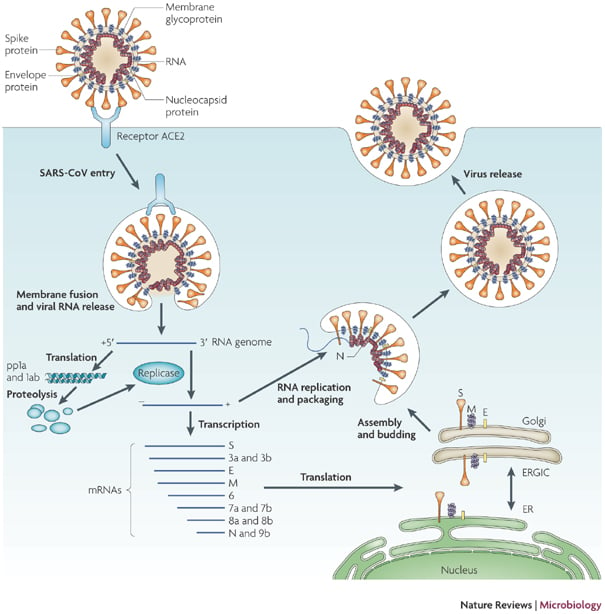
- The first event after uncoating is translation of the viral genomic RNA to produce a virus-specific RNA-dependent RNA polymerase.
- The viral polymerase transcribes a full length complementary (minus-strand) RNA that serves as the template for a nested set of five to seven subgenomic mRNAs.
- Only the 5′ terminal gene sequence of each mRNA is translated.
- Newly synthesized genomic RNA molecules interact in the cytoplasm with the nucleocapsid protein to form helical nucleocapsids.
- The nucleocapsids bud through membranes of the rough endoplasmic reticulum and the Golgi apparatus in areas that contain the viral glycoproteins.
- Mature virions may then be transported in vesicles to the cell periphery for exit.
Pathogenesis of SARS-CoV
- SARS CoV targets type 1 and type 2 alveolar epithelial cells of the lung and also differentiated bronchial epithelial cells.
- The desquamation of alveolar epithelial cells leads to hyaline membrane formation within the alveoli and diffuse alveolar damage, the histological hallmark of acute respiratory distress syndrome (ARDS).
- Patients with SARS have elevated levels of pro-inflammatory cytokines (IL-6, IL-12) and chemokines (IL-8, CCL-2, CXCL10) in the plasma.
- Viral load in the upper respiratory tract peaks around days 7–10 of the disease and falls thereafter, while the lung pathology appears to progress through the second week of illness, suggesting that the lung pathology continues to be driven by mechanisms other than viral replication alone.
- The SARS CoV also infects the intestinal epithelium and virus is shed in the faeces.
- The diarrhoea associated with SARS infection may be related in part to direct infection of the intestinal tract.
Clinical Manifestations of SARS
- The incubation period of SARS is estimated to be 2–14 days.

- The symptoms include
- fever over 100.4°F
- dry cough
- sore throat
- problems breathing, including shortness of breath
- headache
- body aches
- loss of appetite
- malaise
- night sweats and chills
- confusion
- rash
- diarrhea
- SARS presented as a severe progressive ‘atypical pneumonia.
- SARS involves a cytokine storm, with elevated levels of multiple chemokines and cytokines in the peripheral circulation for about 2 weeks.
- Some patients progressed to increasing tachypnoea, oxygen desaturation and respiratory distress syndrome.
- Moderate liver dysfunction and marked lymphopenia is seen.
- Complication include liver failure and heart failure.
Lab Diagnosis of SARS-CoV
- A chest X-ray or CT scan may also reveal signs of pneumonia characteristic of SARS.
- Electron microscopy (EM) on respiratory specimens including lung tissue obtained at open-lung biopsy or autopsy.
- SARS viruses have been recovered from oropharyngeal specimens using Vero monkey kidney cells.
- Detection of the viral RNA genome in respiratory and stool samples by RT-PCR.
- Electron microscopy of negatively stained stool specimens is useful for the detection of enteric coronaviruses
- Serology using enzyme-linked immunosorbent assay (ELISA) can be used to evaluate acute and convalescent sera.
- Complement fixation, immunofluorescence or virus neutralization tests have been used for serological diagnosis.
Treatment of SARS
- Mechanical ventilation and critical care treatment may be necessary during the illness.
- A number of therapeutic options including ribavirin, interferon alpha, lopinavir/ ritonavir, and nucleoside analogue, protease inhibitor combination therapy were used.
- However, there is not yet enough evidence to prove that these treatments are effective.
- Researchers are currently working on a vaccine for SARS, but there have been no human trials for any potential vaccine.
Prevention and control of SARS
- Reduce the contact with SARS infected individuals.
- Handwashing and personal hygiene
- Covering mouth and nose while sneezing to avoid droplet infection.
- Avoid sharing food and drinks from an infected person.
- Disinfection of the working space and surroundings.
