- Recombinant DNA technology refers to the joining together of DNA molecules from two different species that are inserted into a host organism to produce new genetic combinations that are of value to science, medicine, agriculture, and industry.
- Recombinant DNA (rDNA), on the other hand is the general name for a piece of DNA that has been created by the combination of at least two strands.
- They are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) to bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome.
- Recombinant DNA in a living organism was first achieved in 1973 by Herbert Boyer, of the University of California at San Francisco, and Stanley Cohen, at Stanford University, who used E. coli restriction enzymes to insert foreign DNA into plasmids.
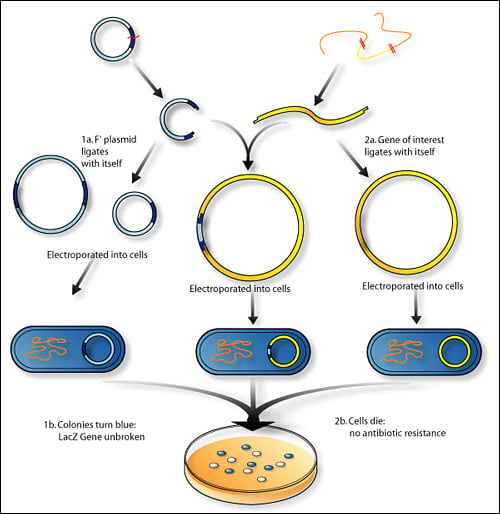
Steps of Genetic Recombination Technology
-
Isolation of Genetic Material
- The first step in rDNA technology is to isolate the desired DNA in its pure form i.e. free from other macromolecules.
- Since DNA exists within the cell membrane along with other macromolecules such as RNA, polysaccharides, proteins, and lipids, it must be separated and purified which involves enzymes such as lysozymes, cellulase, chitinase, ribonuclease, proteases etc.
- Other macromolecules are removable with other enzymes or treatments. Ultimately, the addition of ethanol causes the DNA to precipitate out as fine threads. This is then spooled out to give purified DNA.
Restriction Enzyme Digestion
- Restriction enzymes act as molecular scissors that cut DNA at specific locations. These reactions are called ‘restriction enzyme digestions’.
- They involve the incubation of the purified DNA with the selected restriction enzyme, at conditions optimal for that specific enzyme.
- The technique ‘Agarose Gel Electrophoresis’ reveals the progress of the restriction enzyme digestion.
- This technique involves running out the DNA on an agarose gel. On the application of current, the negatively charged DNA travels to the positive electrode and is separated out based on size. This allows separating and cutting out the digested DNA fragments.
- The vector DNA is also processed using the same procedure.
Amplification Using PCR
- Polymerase Chain Reaction or PCR is a method of making multiple copies of a DNA sequence using the enzyme – DNA polymerase in vitro.
- It helps to amplify a single copy or a few copies of DNA into thousands to millions of copies.
- PCR reactions are run on ‘thermal cyclers’ using the following components:
- Template – DNA to be amplified
- Primers – small, chemically synthesized oligonucleotides that are complementary to a region of the DNA.
- Enzyme – DNA polymerase
- Nucleotides – needed to extend the primers by the enzyme.
- The cut fragments of DNA can be amplified using PCR and then ligated with the cut vector.
- Ligation of DNA Molecules
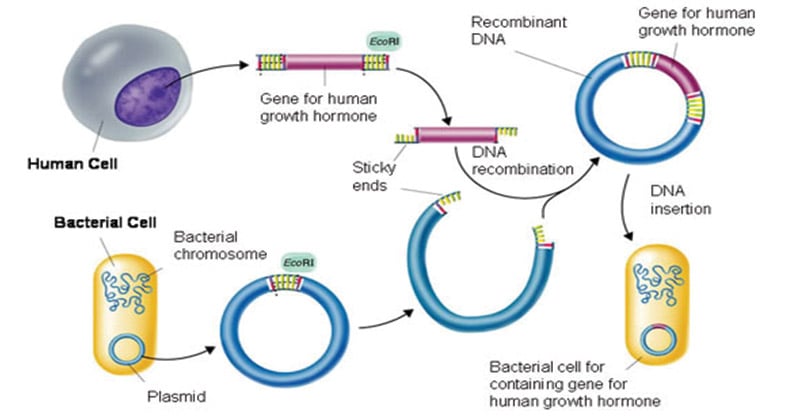
- The purified DNA and the vector of interest are cut with the same restriction enzyme.
- This gives us the cut fragment of DNA and the cut vector, that is now open.
- The process of joining these two pieces together using the enzyme ‘DNA ligase’ is ‘ligation’.
- The resulting DNA molecule is a hybrid of two DNA molecules – the interest molecule and the vector. In the terminology of genetics this intermixing of different DNA strands is called recombination.
- Hence, this new hybrid DNA molecule is also called a recombinant DNA molecule and the technology is referred to as the recombinant DNA technology.
- Insertion of Recombinant DNA Into Host
- In this step, the recombinant DNA is introduced into a recipient host cell mostly, a bacterial cell. This process is ‘Transformation’.
- Bacterial cells do not accept foreign DNA easily. Therefore, they are treated to make them ‘competent’ to accept new DNA. The processes used may be thermal shock, Ca++ ion treatment, electroporation etc.
- Isolation of Recombinant Cells
- The transformation process generates a mixed population of transformed and non-trans- formed host cells.
- The selection process involves filtering the transformed host cells only.
- For isolation of recombinant cell from non-recombinant cell, marker gene of plasmid vector is employed.
- For examples, PBR322 plasmid vector contains different marker gene (Ampicillin resistant gene and Tetracycline resistant gene. When pst1 RE is used it knock out Ampicillin resistant gene from the plasmid, so that the recombinant cell become sensitive to Ampicillin.
Interesting Science Videos
Application of Recombinant DNA technology
- Recombinant DNA is widely used in biotechnology, medicine and research.
- The most common application of recombinant DNA is in basic research, in which the technology is important to most current work in the biological and biomedical sciences.
- Recombinant DNA is used to identify, map and sequence genes, and to determine their function.
- Recombinant proteins are widely used as reagents in laboratory experiments and to generate antibody probes for examining protein synthesis within cells and organisms.
- Many additional practical applications of recombinant DNA are found in industry, food production, human and veterinary medicine, agriculture, and bioengineering.
- DNA technology is also used to detect the presence of HIV in a person.
- Application of recombinant DNA technology in Agriculture – For example, manufacture of Bt-Cotton to protect the plant against ball worms.
- Application of medicines – Insulin production by DNA recombinant technology is a classic example.
- Gene Therapy – It is used as an attempt to correct the gene defects which give rise to heredity diseases.
- Clinical diagnosis – ELISA is an example where the application of recombinant DNA is possible.
Limitations of Recombinant DNA technology
- Destruction of native species in the environment the genetically modified species are introduced in.
- Resilient plants can theoretically give rise to resilient weeds which can be difficult to control.
- Cross contamination and migration of proprietary DNA between organisms.
- Recombinant organisms contaminating the natural environment.
- The recombinant organisms are population of clones, vulnerable in exact same ways. A single disease or pest can wipe out the entire population quickly.
- Creation of superbug is hypothesized.
- Ethical concern about humans trying to play God and mess with the nature’s way of selection. It is exaggerated by the fear of unknown of what all can be created using the technology and how is it going to impact the civilization.
- Such a system might lead to people having their genetic information stolen and used without permission.
- Many people worry about the safety of modifying food and medicines using recombinant DNA technology.
References
- Verma, P. S., & Agrawal, V. K. (2006). Cell Biology, Genetics, Molecular Biology, Evolution & Ecology (1 ed.). S .Chand and company Ltd.
- Klug, W. S., & Cummings, M. R. (2003). Concepts of genetics. Upper Saddle River, N.J: Prentice Hall.
- https://byjus.com/biology/recombinant-dna-technology/
- https://en.wikipedia.org/wiki/Recombinant_DNA
- https://www.britannica.com/science/recombinant-DNA-technology
- https://www.quora.com/What-are-the-advantages-and-disadvantages-of-recombinant-DNA.

These are fascinating lectures, informations, and summaries, since I am a retired doctor, I am spending my time reading only, I have nothing to do, congratulation for this effort and thank you for all of you, please accept my respect to all of you
RASHID ELTAYEB