- Deoxyribonucleic acid (DNA) extraction is the process by which DNA is separated from proteins, membranes, and other cellular material contained in the cell from which it is recovered.
- The simplest cells, such as bacteria cells, are prokaryotes. These prokaryotes comprise a lipid bilayer outer membrane and a cytoplasm containing a circular chromosome, proteins inorganic salts and metal ions, sugar molecules, and other elements of cell machinery.
- A large number of different protocols for the efficient isolation of highly purified DNA from both eukaryotic and prokaryotic cells is extant.
- Many of the available DNA extraction procedures have common elements. Indeed, the extraction of DNA generally follows three basic steps:
- Lyse (break open) the cells.
- Separate the DNA from the other cell components.
- Isolate the DNA.
Interesting Science Videos
Phenol-chloroform extraction of DNA
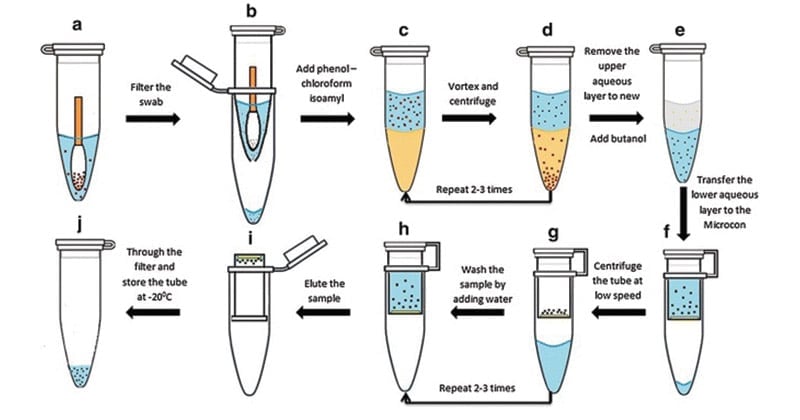
A phenol-chloroform extraction is a liquid-liquid extraction. A liquid-liquid extraction is a method that separates mixtures of molecules based on the differential solubilities of the individual molecules in two different immiscible liquids. Liquid-liquid extractions are widely used to isolate RNA, DNA, or proteins.The extraction of nucleic acids involves adding an equal volume of phenol-chloroform to an aqueous solution of lysed cells or homogenized tissue, mixing the two phases, and allowing the phases to separate by centrifugation. Centrifugation of the mixture yields two phases: the lower organic phase and the upper aqueous phase.
Principle
- The organism to be used should be grown in a favorable medium at an optimal temperature, and should be harvested in late log to early stationary phase for maximum yield.
- The genomic DNA isolation needs to separate total DNA from RNA, protein, lipid, etc.
- Initially the cell membranes must be disrupted in order to release the DNA in the extraction buffer. SDS (sodium dodecyl sulphate) is used to disrupt the cell membrane.
- Once cell is disrupted, the endogenous nucleases tend to cause extensive hydrolysis. DNA can be protected from endogenous nucleases by chelating Mg2++ ions using EDTA. Mg2++ ion is considered as a necessary cofactor for action of most of the nucleases.
- Nucleoprotein interactions are disrupted with SDS, phenol or proteinase K.
- Proteinase enzyme is used to degrade the proteins in the disrupted cell soup.
- Phenol and chloroform are used to denature and separate proteins from DNA. Chloroform is also a protein denaturant, which stabilizes the rather unstable boundary between an aqueous phase and pure phenol layer.
- The denatured proteins form a layer at the interface between the aqueous and the organic phases which are removed by centrifugation.
- DNA released from disrupted cells is precipitated by cold absolute ethanol or isopropanol.
Materials and Reagents
- Tris base
- Proteinase K
- Phenolchloroform (1:1)
- 200 proof ethanol
- RNAase
- Ethanol
- SDS
- EDTA
- Tryptone
- Yeast extract
- NaCl
- LB medium
- TE buffer
- Lysis buffer
Equipment
- Tabletop centrifuge
- 1.5 ml Eppendorf tube
- Incubator
Procedure
- Transfer 1.5 ml of the overnight E. coli culture (grown in LB medium) to a 1.5 ml Eppendorf tube and centrifuge at max speed for 1min to pellet the cells.
- Discard the supernatant without disturbing the cell pellet.
- Resuspend the cell pellet in 600 μl lysis buffer and vortex to completely resuspend cell pellet.
- Incubate 1 h at 37 °C.
- Add an equal volume of phenol/chloroform and mix well by inverting the tube until the phases are completely mixed.
- Spin at max speed for 5 min at RT (all spins are performed at RT, unless indicated otherwise). There is a white layer (protein layer) in the aqueous: phenol/chloroform interface.
- Carefully transfer the upper aqueous phase to a new tube by using 1 ml pipetman (to avoid sucking the interface, use 1 ml tip with wider mouth-cut 1 ml tip-mouth about ~2 mm shorter).
- Steps 4-6 can be repeated until the white protein layer disappears.
- To remove phenol, add an equal volume of chloroform to the aqueous layer. Again, mix well by inverting the tube.
- Spin at max speed for 5 min.
- Remove aqueous layer to new tube.
- To precipitate the DNA, add 2.5 or 3 volume of cold 200 proof ethanol (store ethanol at -20 °C freezer) and mix gently (DNA precipitation can be visible).
Note: DNA precipitation may simply diffuse, which is normal. Keep the tube at -20 degree for at least 30 min (the longer the better) and then spin it down (see Steps 15-16). You should see DNA pellet. It looks transparency when it is wet and turns to white when it becomes dry. - Incubate the tube at -20 °C for 30 min or more.
- Spin at max speed for 15 min at 4 °C.
- Discard the supernatant and rinse the DNA pellet with 1 ml 70% ethanol (stored at RT).
- Spin at max speed for 2 min. Carefully discard the supernatant and air-dry the DNA pellet (tilt the tube a little bit on paper towel). To be faster, dry the tube at 37 °C incubator.
- Resuspend DNA in TE buffer.
Note: Large amounts of RNA will be present in the DNA sample. So, for subsequent reactions, for example, to digest plasmid DNA, add 1-5 μl (1 mg ml-1) RNAase to the digestion solution to completely remove RNA. Or, add RNAase directly to lysis buffer with a final concentration of 1 mg ml-1. - Isolated Gemonic DNA can be subjected to agarose gel electrophoresis.
Expected Results
- White strands of DNA precipitate.
- On gel, bands with smear patterns from high to low molecular weight range can be seen.
- Most of DNA fragments accumulated at high molecular weight: Not degraded.
- Most of DNA fragments small: DNA might have got degraded.
References
- He, F. (2011). E. coli Genomic DNA Extraction. Bio-protocol Bio101: e97. DOI: 21769/BioProtoc.97.
- Maniatis T., E.F. Fritsch, and J. Sambrook (1982). Molecular Cloning A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Springs Harbor, NY.
- http://www.methodquarterly.com/2014/11/protocol-dna-extractions/
- https://genome.cshlp.org/content/4/6/368.full.pdf
- http://labcenter.dnalc.org/labs/dnaextraction/pdfs/teacher/Lesson%20Plan%20DNA%20Extraction%20from%20Bacteria.pdf
- https://www.sciencedirect.com/topics/neuroscience/dna-extraction
- http://www.srmuniv.ac.in/sites/default/files/files/BT0210%20%20MOLECULAR%20BIOLOGY%20LABORATORYMANUAL.pdf
- http://physiology.med.cornell.edu/faculty/mason/lab/zumbo/files/PHENOL-CHLOROFORM.pdf
- http://www.protocol-online.org/cgi-bin/prot/view_cache.cgi?ID=2052

Thanks a lot for this protocol. Could you please share a protocol for the lysis buffer? Thanks