Proteomic sequencing is a method that is used to study and characterize proteins in a biological sample. It is a process of identifying the exact order of amino acids in a protein which reveals its structure and function in biological processes.
Proteins are important biological molecules that are involved in many processes including structural roles, metabolic processes, transport, signaling, and regulatory roles so proteomic sequencing is useful for understanding complex cellular processes and disease mechanisms.
Genetic information is passed from DNA to RNA which is then used to produce proteins. So, amino acid sequences can also be decoded from gene sequences but it cannot provide additional information like post-translational modifications, protein folding, and interactions which are important for protein function. High-throughput and direct proteomic methods are needed to get detailed information about proteins. Proteomic sequencing is a direct method for studying protein structure, function, interactions, and modifications within a biological system.
What is Proteomics?
All the proteins expressed in an organism at a given time are known as its proteome and the study of proteome is called proteomics. Proteomics studies the interactions and specific functions of proteins within an organism which helps to understand how they contribute to biological processes and diseases.
In proteomics, there are three different approaches for studying proteins in a sample:
- Top-down Proteomics: In this method, intact proteins in a sample are first separated and directly analyzed. This method is useful for studying post-translational modifications (PTMs) and protein isoforms as it allows direct analysis of intact proteins. However, it faces challenges when analyzing complex mixtures of proteins.
- Bottom-up Proteomics: This method involves digesting proteins into smaller peptides before analyzing them. The peptides are separated and identified using methods like chromatography and mass spectrometry. This method is suitable for identifying and quantifying a large number of proteins as the smaller peptide fragments are easier to analyze but it does not provide information about intact proteins or their modifications.
- Middle-down Proteomics: This is a relatively new approach that lies between top-down and bottom-up proteomics. In this method, proteins are digested to generate larger peptides rather than being fully digested into smaller peptides. This approach provides more detailed analysis of protein structure and post-translational modifications.
Principle of Proteomic Sequencing
Proteomic sequencing involves identifying and characterizing proteins to identify their structure, function, and interactions. The main principle of proteomic sequencing is to separate, identify, and quantify proteins in a sample.
The process of proteomic sequencing begins with the isolation of target protein from a biological sample. It is then broken down into smaller fragments called peptides. These peptides are analyzed using methods like mass spectrometry and Edman degradation which helps determine their sequence. The full protein sequence can be reconstructed by comparing the sequence with known protein databases.
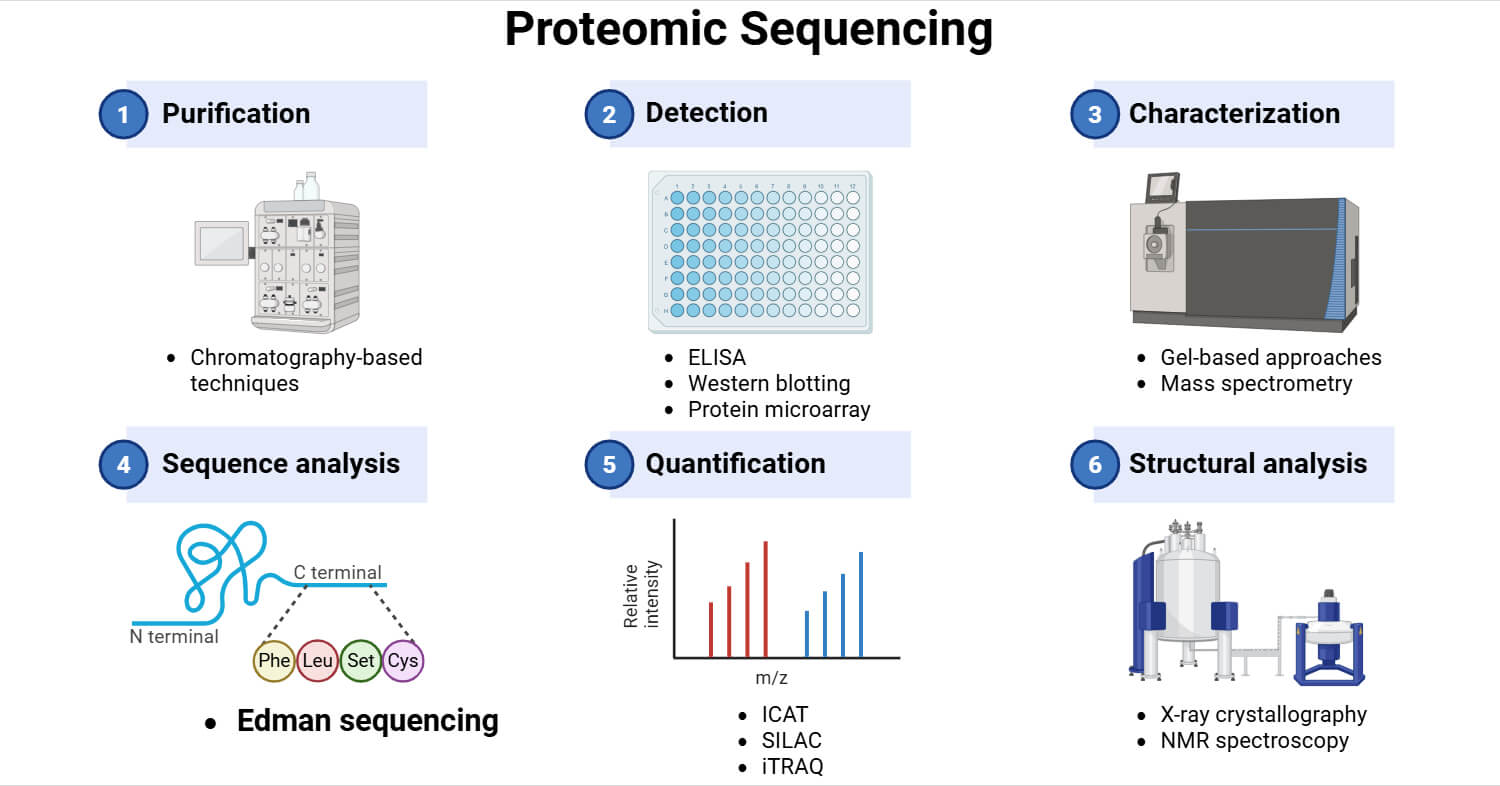
Steps of Proteomic Sequencing
1. Sample Preparation
Initially, the proteins of interest are extracted from biological samples. The extraction process includes the use of organic solvents, detergents, and other methods to lyse the cells and release proteins. Organic solvents and detergents are removed by lyophilization to improve protein detection.
2. Protein Separation
Then, proteins are separated using gel-based or chromatography-based methods. Gel-based methods include 1-Dimensional Electrophoresis (1-DE) and 2-Dimensional Electrophoresis (2DE). 1-DE separates proteins based on molecular weight. 2-DE provides better resolution by separating proteins based on molecular mass and isoelectric point. DIGE is a modification of 2-DE that uses fluorescent dyes to compare protein samples on the same gel. Chromatography-based methods separate proteins from complex mixtures. It includes methods like Ion Exchange Chromatography (IEC), Size Exclusion Chromatography (SEC), Affinity Chromatography, and Liquid Chromatography (LC).
3. Sequencing
After separation and purification of proteins, both ends of the polypeptide are sequenced. Each peptide fragment is sequenced individually using different methods like Edman sequencing and mass spectrometry. Recently, newer methods for proteomic sequencing have been developed such as fluorosequencing, tunnelling current, and nanopore-based protein sequencing.
4. Data Analysis
The raw data generated from sequencing is preprocessed and the data is normalized to correct experimental biases. Then, the peptides are aligned to known proteins using databases and algorithms. This is used to identify proteins, quantify them, and study their interactions and post-translational modifications.
Methods of Proteomic Sequencing
a. Mass Spectrometry (MS)
- MS is a commonly used method in proteomic sequencing that measures the mass-to-charge ratio of peptide ions and determines the chemical structures of proteins.
- In this method, peptides are first ionized using an ion source. The two main ionization methods used in mass spectrometry are electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI).
- Once ionized, peptides are separated and analyzed using different mass analyzers like Time-of-Flight (TOF) and Ion Trap.
- For more detailed sequencing, tandem mass spectrometry (MS/MS) is used. This method involves two stages of mass spectrometry.
- MS can be also be combined with methods like liquid chromatography (LC-MS) to analyze complex mixtures of proteins.
b. Edman Degradation
- It is a method that sequentially removes and identifies amino acids from the N-terminal end of a peptide. It is considered the first protein sequencing method.
- The process begins with a Phenylisothiocyanate (PITC) reaction which reacts with the protein of interest at the N-terminus and forms a stable Phenylthiocarbamyl (PTC) derivative.
- Then, the N-terminal amino acid is cleaved using TFA (trifluoroacetic acid) and identified using chromatographic methods or mass spectrometry. This is repeated to sequence the entire protein.
- It allows sequencing of peptides by keeping the protein intact and provides accurate sequence data but it is less efficient for larger proteins.
c. Nanopore sequencing
- Nanopore sequencing was originally developed for DNA and RNA analysis but advances in nanopore design have made it possible to use this technology to proteomics.
- This method involves passing the proteins through a nanopore and measuring the electrical current as the protein moves through the nanopore. Each amino acid or modification causes a unique disruption in the current which allows the identification of specific residues.
- Motor proteins and electro-osmotic flow can be used to control the speed of peptides which helps overcome challenges associated with the rapid and uneven movement of proteins during sequencing.
- Nanopore sequencing for proteomic is still in the experimental stage but it has potential applications in real-time and high-throughput protein analysis.
d. Fluorosequencing
- This method uses fluorescence to detect and sequence peptides or proteins.
- In this method, the protein is tagged with a fluorescent marker and sequenced. The fluorescent signals from each amino acid are detected and analyzed.
- It can provide detailed sequence information for small peptides but is limited by the need for fluorescent labeling and is not widely used for large-scale sequencing.
e. Tunneling current-based sequencing
- This method involves using a tunneling current to measure changes as peptides pass through a nanopore and provides sequence information.
- The peptide sequence is determined by the electrical signals generated as individual amino acids move through the nanopore.
- It allows high-throughput sequencing but this technology is still under development.
Advantages of Proteomic Sequencing
- Proteomic sequencing helps to identify and quantify proteins in complex biological samples.
- It provides direct information on the proteins present in a sample unlike genome sequencing.
- It can detect low-abundance proteins including biomarkers for diseases.
- It helps to understand signaling pathways and disease mechanisms.
- Proteomics can distinguish protein isoforms and alternative splice variants.
- It can identify chemical modifications which regulate protein function and cannot be predicted by DNA or RNA sequencing.
Limitations of Proteomic Sequencing
- It is difficult to sequence larger or highly modified proteins.
- Sample preparation is often difficult and time-consuming. Proteins must be carefully extracted, purified, and digested before sequencing.
- Large-scale proteomic studies can be expensive especially when using mass spectrometry.
- Edman degradation method is less effective for long or modified proteins.
- It is difficult to fully capture and analyze the wide variety of proteomes.
- It produces large amount of data which needs specialized bioinformatics tools.
Applications of Proteomic Sequencing
- Proteomic sequencing has applications in identifying novel proteins or isoforms.
- It is also used to identify protein biomarkers that can help in disease diagnosis.
- It also has applications in personalized medicine. It can be used to tailor disease treatment to individual patients based on their genetic profiles.
- Proteomic sequencing can analyze post-translational modifications like phosphorylation and glycosylation.
- It can also be useful in drug discovery and development. It helps to identify proteins that are important in diseases.
- Proteomic sequencing is useful in developing biopharmaceuticals, recombinant proteins, and enzymes.
References
- Al-Amrani, S., Al-Jabri, Z., Al-Zaabi, A., Alshekaili, J., & Al-Khabori, M. (2021). Proteomics: Concepts and applications in human medicine. World Journal of Biological Chemistry, 12(5), 57–69. https://doi.org/10.4331/wjbc.v12.i5.57
- An overview of proteomics. (2025, February 19). Retrieved from https://www.abcam.com/en-us/knowledge-center/proteins-and-protein-analysis/proteomics
- Beeton-Kempen, N. (2024, July 5). Proteomics: Principles, techniques and applications. Retrieved from https://www.technologynetworks.com/proteomics/articles/proteomics-principles-techniques-and-applications-343804
- Cui, M., Cheng, C., & Zhang, L. (2022). High-throughput proteomics: a methodological mini-review. Laboratory Investigation, 102(11), 1170–1181. https://doi.org/10.1038/s41374-022-00830-7
- Gobena, S., Admassu, B., Kinde, M. Z., & Gessese, A. T. (2024). Proteomics and its current application in biomedical area: Concise review. The Scientific World JOURNAL, 2024, 1–13. https://doi.org/10.1155/2024/4454744
- Li, Z., Yi, Y., Liu, L., & Wu, H. (2024). One step forward for nanopore protein sequencing. Clinical and Translational Medicine, 14(3). https://doi.org/10.1002/ctm2.1615
- Methods and techniques for protein sequencing – Creative Proteomics. Retrieved from https://www.creative-proteomics.com/resource/methods-and-techniques-for-protein-sequencing.htm
- Protein sequencing: Significance, methods, and applications – Creative Proteomics. Retrieved from https://www.creative-proteomics.com/resource/protein-sequencing-significance-methods-applications.htm
- Ritmejeris, J., Chen, X., & Dekker, C. (2024). Single-molecule protein sequencing with nanopores. Nature Reviews Bioengineering. https://doi.org/10.1038/s44222-024-00260-8
