Interesting Science Videos
What are Protein synthesis inhibitors?
Protein synthesis, a long process that includes different enzymes and structural change in organisms. Some antibacterial classes inhibit bacterial protein synthesis by interfering with the 30s or 50s subunit. Antibiotics target three different steps which include initiation, formation of the 70s, and elongation process of making polypeptides. Antibiotics that inhibit or interferes with bacterial protein synthesis include; Aminoglycosides, Macrolides, Tetracycline, Oxazolidinone, and Chloramphenicol.
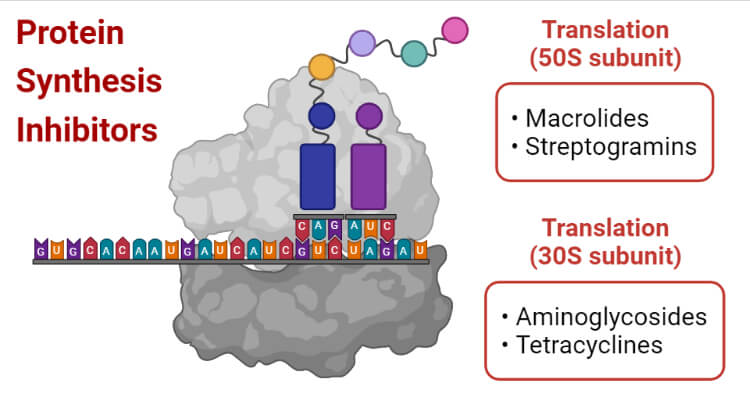
- Aminoglycosides: They consist of different amino acids with different functions and are hydrophilic sugars. As aminoglycosides are polycationic they have a high binding affinity for nucleic acids. Different antibiotics of aminoglycosides bind to different sites on rRNA and have different affinities but some antibiotics bind to the same site in more than one confirmation. Like, paromomycin binds to A sites of rRNA. Aminoglycosides only interact with the nucleic acid bases but not with the phosphate backbone. Electronic and Stearic constraints should be satisfied in case of not binding with RNA.
- Tetracycline: They consist of A, B, C, and D nucleus rings in which several functional groups are attached. They interact with the ribosome and show reversible bacteriostatic properties by inhibiting bacterial protein synthesis.
- Chloramphenicol: Before chloramphenicol was referred to as chloromycetin which was originally isolated from Streptomyces venezuelae. As it broad-spectrum antibiotic it is used to treat both Gram-positive and Gram-negative infections and it is very stable. They are protein synthesis inhibitors that prevent the bacteria from peptide chain elongation.
- Macrolides: Macrolides that is used in treating Gram-positive infections is made up of 14, 15 or 16 membered lactone ring. Macrolides are protein synthesis inhibitors as it binds to ribosomal subunit in peptidyl transferase center and causing cell lysis by inhibiting protein synthesis.
- Oxazolidinones: Oxazolidinones are a newer class of antibiotics and linezolid is the first commercially available oxazolidinone. They are considered as the last resort drug which is used in treating Vancomycin-resistant infections and other infections caused by Gram-positive bacteria. Oxazolidinone acts by inhibiting initiation complex, binding to P-site, and translocation of peptidyl-tRNA from A-site to P-site.
Mechanism of action of Protein synthesis inhibitors
Protein synthesis inhibitors usually act at the ribosomal level in the translation process of protein synthesis that includes initiation, elongation, and termination. In both prokaryotes and eukaryotes, ribosomes are the main site for protein synthesis. Mainly, tRNA binds to three sites of mRNA complex; A-site or aminoacyl site, Peptidyl site or P-site, and E site or Exit site.
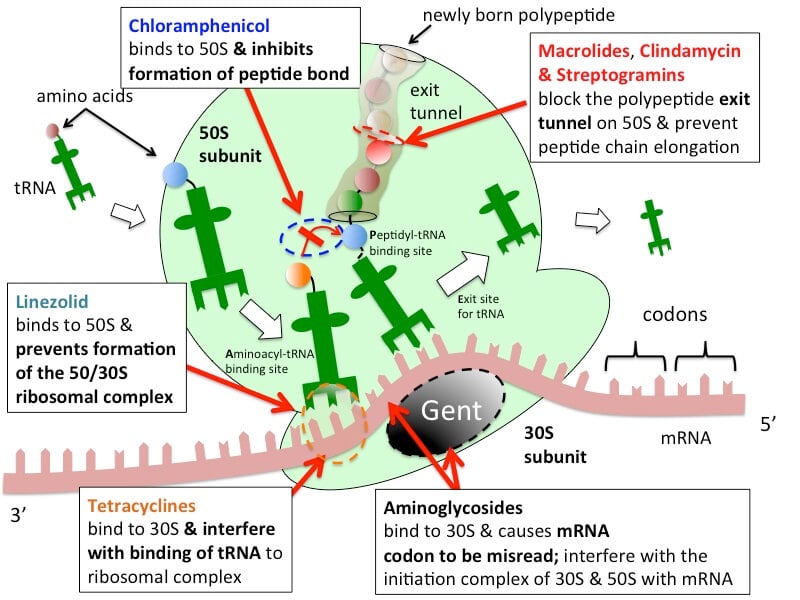
Figure: Sites of action of antibiotics that interfere with bacterial protein synthesis. Image Source: Tulane University.
Mechanism of action of the different class of antibiotics on protein synthesis are described below:
Aminoglycosides: When aminoglycosides get their entry into the cytoplasm, inhibition of bacterial protein synthesis and mistranslation of protein occurs. They bind to the A-site of 16s rRNA with high affinity. Mistranslation occurs after the codon misreading on delivery of aminoacyl tRNA. This causes incorrect amino acids to change into polypeptides that result in cell damage. Some aminoglycosides inhibit protein synthesis by blocking in elongation stage of translation.
Macrolides: For different modes were described in the mode of action shown by macrolides in inhibition of bacterial protein synthesis.
- During the early phase of translation progression of peptide chain inhibition occurs.
- Peptidyl tRNA dissociation occurs from the ribosome.
- Hindrance in peptide bond formation
- Intervention with 50s subunit.
In presence of some amino acid, macrolides were found by the ribosome which results in inhibition. And on some extent macrolides cause frameshifting causing rough bacterial protein synthesis.
Chloramphenicol: Chloramphenicol affects the elongation steps in bacterial protein synthesis by inhibiting the peptidyl transferase activity. Chloramphenicol binds to A2451 and A2452 of ribosomal units that prevent peptide bond formation resulting in inhibition of protein synthesis.
Tetracycline: Tetracyclines are lipophilic molecules so they can cross the cell membrane. They are bacteriostatic. They bind to the 30s ribosomal unit by preventing attachment of transfer RNA which interferes with amino acids to form proteins and inhibit bacterial protein synthesis.
Oxazolidinone: Oxazolidinones inhibit the formation of the initiation complex which is composed of mRNA, GTP, and other initiation factors. They bind to the 50s ribosomal subunit. Inhibition of formation of 70s subunit is by the after effect shown by binding to 50s subunit complex. In the case of 70s formation translocation of peptide chain from P-site to A-site is inhibited which results in inhibition of protein synthesis.
Examples of Protein synthesis inhibitors
- Oxazolidinone: Linezolid
- Tetracycline: Doxycycline
- Macrolide: Azithromycin, Erythromycin
- Chloramphenicol: Chloromycetin
- Aminoglycosides: Amikacin, Kanamycin
Mechanism of Resistance
- Tetracyclines: Efflux pump, a mechanism that removes active antibiotics from the cell causing antibiotics to pump from the cell because of trans membrane proteins. This mechanism exports the molecule with magnesium ions causing a lower concentration of antibacterials within the cells. Cytoplasmic proteins that protect the ribosome from tetracycline are also responsible for causing tetracycline resistance.
- Chloramphenicol: Efflux mechanisms, the mutation in the target site, permeability barriers, and inactivation of an enzyme phosphotransferases by acetylation. The loss of outer membrane protein plays important role in resistance. Acetylation occurs through different types of chloramphenicol acetyltransferases that are capable of inactivating chloramphenicol. catA and catB are genes that code chloramphenicol acetyltransferases.
- Aminoglycosides: Resistance to aminoglycosides is caused by the methylation of 16s rRNA. The common mechanism of aminoglycosides includes modification in structures by different enzymes. Three classes of enzymes are aminoglycoside phosphotransferases, aminoglycoside nucleotidyltransferases, and aminoglycoside acetyltransferases. Modification in structures causes ability of antibiotics ability due to the steric or electrostatic interactions.
- Macrolides: Macrolide resistance is mainly due to the transcription modification of 23s rRNA or mutation caused by erm genes. Modification in the binding site of macrolide is due to the demethylation of A2058 residue in 23s rRNA by erm- methyltransferases. These erm genes cause resistance in pathogens by horizontal gene transfer. Steric hindrance decreases drug affinity by the dimethylation of A2058 residues which results in cell resistance to antibiotics of macrolides.
- Oxazolidonones: resistance to oxazolidinone is due to the target modification and due to cfr (chloramphenicol-florfenicol resistant) genes. And also due to mutation caused by Guanine to Uracil substitution of 23s rRNA at A2058 position. Cfr genes are both plasmid and as well as chromosomal mediated.
References
- Bozdogan B and Appelbaum, PC (2019). Oxazolidinones : Activity, mode of action, and mechanism of resistance Oxazolidinones : activity, mode of action, and mechanism of resistance. (March 2004). https://doi.org/10.1016/j.ijantimicag.2003.11.003
- Dinos G P, Athanassopoulos C M, Missiri D A, Giannopoulou P C, Vlachogiannis I A, Papadopoulos, GE, Kalpaxis, D. L. (n.d.). Chloramphenicol Derivatives as Antibacterial and Anticancer Agents : Historic Problems and Current Solutions. https://doi.org/10.3390/antibiotics5020020
- Epand R M, Walker C, Epand R F, Magarvey N A and Hilpert K (2016). Biochimica et Biophysica Acta Molecular mechanisms of membrane-targeting antibiotics. BBA – Biomembranes, 1858(5): 980–987. https://doi.org/10.1016/j.bbamem.2015.10.018
- Fyfe C, Grossman T H., Kerstein K and Sutcliffe J (2016). Resistance to Macrolide Antibiotics in Public Health Pathogens. 1–38.
- From, https://www.osmosis.org/learn/Protein_synthesis_inhibitors:_Aminoglycosides.
- From, https://www.frontiersin.org/articles/10.3389/fmicb.2011.00203/full
- Gaynor M and Mankin, A S (2003). Macrolide Antibiotics : Binding Site, Mechanism of Action, Resistance. 1–12.
- Kotra LP, Haddad, J and Mobashery S (2000). MINIREVIEW Aminoglycosides : Perspectives on Mechanisms of Action and Resistance and Strategies to Counter Resistance. 44(12): 3249–3256.
- Nguyen F, Starosta A L, Arenz S., Sohmen, D and Dönhöfer A. (2014). Tetracycline antibiotics and resistance mechanisms. (February). https://doi.org/10.1515/hsz-2013-0292
- Profile SEE (2017). Chloramphenicol Antibiotics. (December). https://doi.org/10.13140/RG.2.2.16151.88484
- Roberts C. (1996). Tetracycline resistance determinants : mechanisms of action , regulation of expression , genetic mobility , and distribution. 19: 1–24.
- Schwarz S, Kehrenberg, C and Cloeckaert A (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. 28: 519–542. https://doi.org/10.1016/j.femsre.2004.04.001
- Singh D (n.d.). Inhibitors of Protein Synthesis.
- Version W (2006). Inhibition of Protein Synthesis by Antibiotics. (13): 37–40.
