Interesting Science Videos
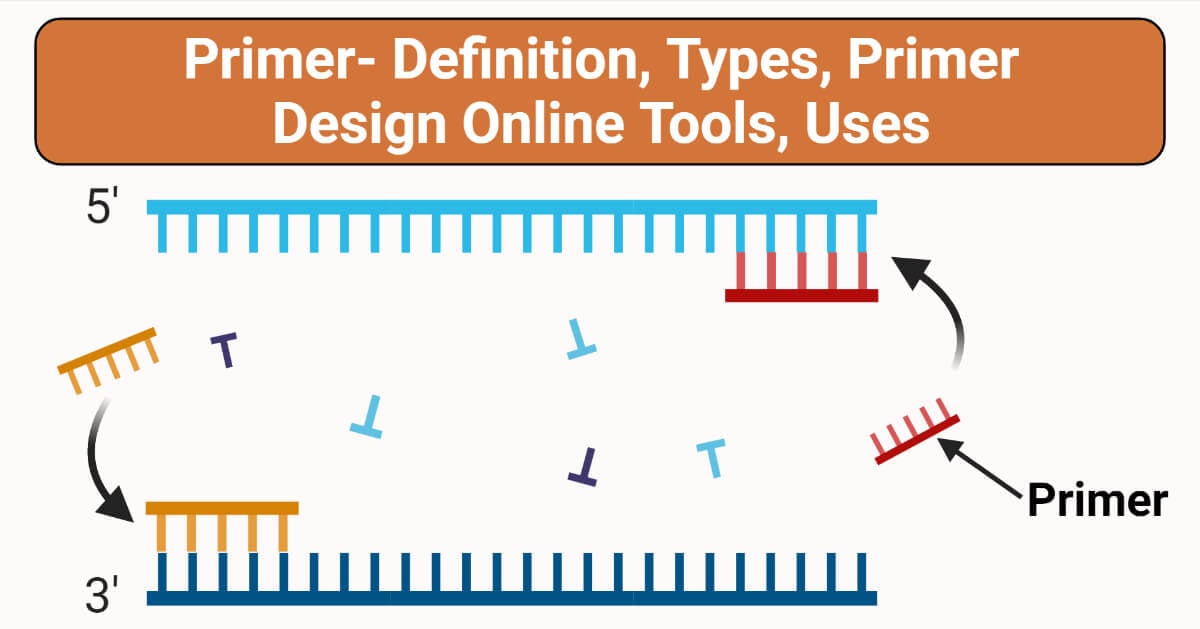
What is Primer?
Primer is a short stretch of sequence that serves as an initiation point for DNA synthesis. There can be a set of primers (forward and reverse) with a sequence complementary to the template DNA -a point of initiation synthesis.
The main objective of the primer is synthesizing DNA with a free terminal end and initiation point of polymerase. A pair of primers one at the template strand while the other at the complementary strand binds on the opposite ends of the sequence being designed, likewise, the 3’ corresponds to the template strand for the process of elongation.
The forward primer runs in 3’-5’ while the reverse primer runs in 5’-3’. However process of elongation results in two new strands of ds DNA.
Caution: The formation of dimers occurs when the template strand lies with the complementary strand which is an unfavorable condition.
Forward and Reverse primers don’t follow the complementarity rule, rather a forward primer binds to one end of one target at 5’ P while the other end of 5’ P occupies reverse primer.
DNA primers were used most widely in PCR, unlike replication, due to:
- The concept of stability in DNA primers as compared to more than RNA primers.
- Since it is the unidirectional mode of polymerization, RNA primers cannot be removed after the end of the reaction.
- DNA primers can be synthesized easily
- However, the majority of the amplification is made from DNA, it is recommended to use specific nucleotide primers only.
Note: Primers participate in the replication process and have the ability to remove RNA primers by the end of the replication process while DNA primers are not.
Types of Primers (DNA primers vs RNA primers)
Solely living organisms utilize RNA primers while invitro involves DNA primers. However, DNA primers are much preferred due to varied reasons such as stability, easy storage, fewer enzymes required to initiate synthesis.
The comparison between DNA and RNA primers is listed below.
DNA Primers: Invitro: PCR amplification, DNA sequencing.
RNA Primers: In vivo: DNA replication, cloning.
Reaction
DNA primers: The process of amplification is temperature dependent with fewer proteins.
RNA primers: The replication process is a catalytic reaction in an enzyme-dependent manner with several proteins.
Length
DNA primers: 18- 24 base pairs.
RNA primers: 10- 20 base pairs.
Synthesis: DNA primers are synthesized chemically while RNA primers require Primase enzyme.
DNA primers are long-lived and more stable while RNA primers are short-lived and are more reactive although, DNA polymerase begins the addition of nucleotides to the reactive 3’ (OH) end of existing nucleic acid, along with the elongation and replication of the parent strand.
Primer Designing
Now, let’s discuss the essential factors for Primer designing: Parameters such as length of the primer, melting point, Primer annealing
Primer length: Oligonucleotides between 18-24 are said to be quiet enough and advantageous so that short primers would bind easily to the template at the annealing temperature.
Melting temperature (52°C-56°C) The GC results of the sequence gives a fair indication of the primer Tm. However, the difference of the primer should not be less than 2°C.
Primer Annealing (Ta): The high Ta results in low PCR product with insufficient primer-template hybridization, while too low Ta will lead to non-specific PCR products caused as a result of a high number of base pairs mismatches.
Ta= 0.3*Tm (primer) +0.7 (product) – 14.9, Tm (primer)
Melting Temperature of the Primer:
Tm (primer)- It measures the least stable primer-template pair.
Tm (product)- It measures the melting temperature of the PCR product.
The modified step annealing can be performed using gradient PCR where temperature can be set to bind primers.
Primer GC content and Clamp: Gene sequencing Primers must possess GC content between 40-60%, with the 3’ end, by with 2 GC bases- GC clamp. However, GC bp with 3 H bonds that are stronger than AT bonds with 2 bonds with the high stability of the primer along with the improvement and specificity of the primer binding.
Setting Restriction Enzyme(RE) Cut Sites: The enzyme called leader sequence permits the higher efficiency for cutting enzymes by adding 3-5 bases to the 5’ end of the total cut site in our target primer.
End Stability: The maximum G allows the binding of 3-5 least bases with the 3’ end. However, a stable 3’ end can reduce false priming.
Caution for designing PCR Primers
The primers can be formed as following types:
- Hairpins: The loop structure formed by the intramolecular interactions within the primer which optimally 3’ end with -2kcal/m and internal hairpin with -3kcal/m can be tolerated.
- Dimers: A structure forming ds DNA by intermolecular interactions between 2 primers. Likewise, if the interaction formed between 2 homologous or the same sense of primer, – called as self-dimers while the opposite primers are called as cross dimers.
- Repeats & Run: The consecutive occurrence of dinucleotide runs in the continuous stretch of a single nucleotide is considered the most important property. The maximum no. of repeats and runs was of 4 dinucleotides and 4 base pairs.
- Primer- Template Cross Homology: Primers should be designed in such a way that no homology within the template is been noticed other than the target site which resulted in non-specific binding and amplification. This can be categorized into 2 types: a) Intra-primer homology: The complementary bases within the same pair in the region of more than 3 bases can cause intramolecular bonding b)Inter-primer homology: Forward and reverse primers with complementary sequences are responsible for intermolecular bonding.
Analyzing primer dimer formation is the primary important caution to be taken care of. However, it involves the determination of Gibbs free energy which aids to be the one. Although 5’ end was found to be more reliable than 3’ end.
Best Primer design online tools
- Primer designing tool (nih.gov)
- Primer3 Input
- Primer3Plus (bioinformatics. NL)
- PrimerQuest – design qPCR assays | IDT (idtdna.com)
- PerlPrimer (sourceforge.net)
- Primer Design with Oligo Primer Analysis Software v. 7
- Real-Time PCR Primer Design – Real-Time PCR Probe Design – GenScript
- www.autoprime.de
Primer design Protocol/ steps/Process
Golden Gate Cloning Method of Primer Designing
This method demands the use of a vector assembly (plasmid) into a single construct with one or multiple DNA fragments. The PCR primers overlap to form restriction sites with adjacent DNA fragments and are designed, however, Type 2S enzyme, along with DNA ligases of the fragments for a directional assembly. Likewise, this method exploits the use of type 2 class of restriction sites, i.e. cut outside of their restriction sites through non-palindromic sticky end overhangs. This method exploits multiple fragments of DNA by using a combination of overhang sequences on their insert fragments for easy annealing with the adjacent ones. However, type 2S restriction sites permit for golden gate assembly thus leaving no restriction sites after cloning.
Primer Design using Gibson Cloning Method
This method relies on recombination despite restriction digestion and ligation for the generation of plasmids. It allows the production of identical homologous sequences overlapping at the ends a^ 20-40 bp long on each side of the linearized vector for both the target DNA fragments. This combines an identical overlap which is cleaved by exonuclease so that complementary part overhangs to it in assembly. In short, this can be defined as a method that considers two pieces of homologs DNA together with overlapping ends of DNA which fuses with them.
Validation of Primers and Probes in qPCR
Once the designing of qPCR primers and probes has been done using available tools, insilico validation is to be performed by BLAST (insilico validation) for the confirmation of targeted gene sequences specificity. The algorithm of BLAST carries out sequence- similarity search against several databases with a set of gapped alignments of links to full database records (Raymaekers M et al, 2009). The query coverage and the maximum identity should be 100%. However, the BLAST program reports a statistical significance, called “expectation value”(E – value) for each alignment which is an indicator for finding the match by chance. E – values ≤ 0.01 convey the homologous sequences (Altschul et al., 1990)( Karlin et al., 1990). E- value measures for assessing potential biological relationships (Raymaekers M et al, 2009). Despite the fact that Insilco tools provide valuable feedback, the specificity of the qPCR assay using the designed primers and probes has to be validated empirically with direct experimental evidence (Bustin et al., 2009).
The specificity for a qPCR product can be affected by the presence of non-specific amplification and can be checked by analyzing the melting curves, also called dissociation curves, generating those qPCR protocols based on ds DNA binding dyes including SYBR green, since they bind to primer-dimer and other reaction artifacts producing a fluorescent signal (Holland et al., 1991) (Heid CA et al., 1996). The melting curves can be carried out in all reported software programs for performing qPCR after amplification (Pfaffl MW, 2004).
Applications of Primer Design
Apart from the PCR, DNA sequencing primers combine with restriction cloning, as well as other DNA new assemblies such as Gibson DNA assembly methods together with Golden Gate method.
Limitations
Gibbs’s free energy plays a very pivot role in primer designing. This is because of the spontaneous reaction at constant temperature and pressure. Thereby, higher G denotes(greater than 0, or positive G) implies an enthalpy to form while secondary structures take low spontaneous reaction with lower G value. The very negative G indicates the affinity to form a structure to linear form with the release of heat in the reverse back manner thus, being more a stable secondary structure(larger negative G values) should be avoided.
References
- Raymaekers M, Smets R, Maes B et al (2009) Checklist for optimization and validation of realtime PCR assays. J Clin Lab Anal 23:145–151
- Karlin S, Altschul SF (1990) Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A 87:2264–2268
- Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410
- Bustin SA, Benes V, Garson JA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622
- Pfaffl MW (2004) Quantifi cation strategies in real-time PCR. In: Bustin SA (ed) A-Z of Quantitative PCR (IUL Biotechnology, No. 5). International University Line (IUL), San Diego, CA, pp 87–112
- Holland PM, Abramson RD, Watson R et al (1991) Detection of specifi c polymerase chain reaction product by utilizing the 50–30 exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A 88: 7276–7280
- Heid CA, Stevens J, Livak KJ et al (1996) Real time quantitative PCR. Genome Res 6: 986–994
