A nucleotide is a pentose sugar linked to a nitrogenous base and a phosphate molecule. Nucleotides are the building blocks of DNA.
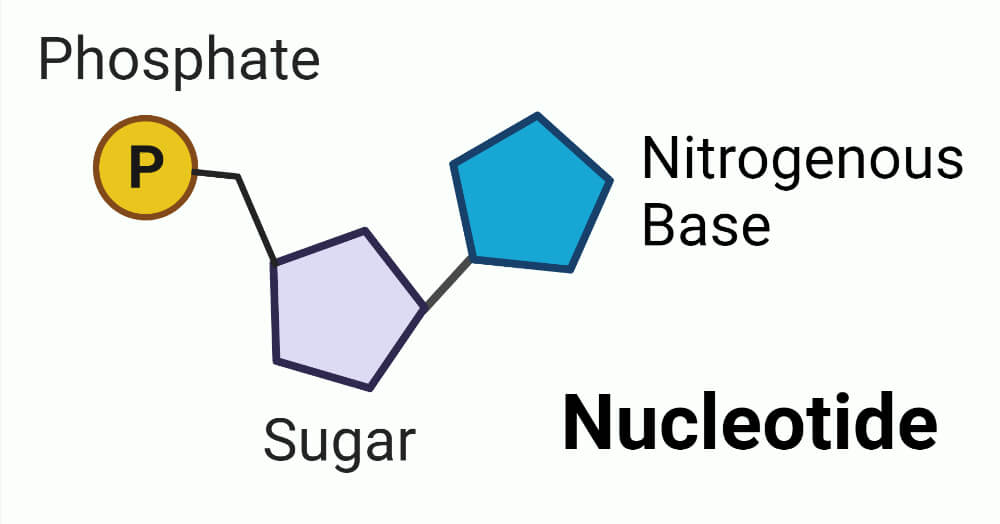
- The nitrogenous bases are derived from two-parent compounds – purines and pyrimidines.
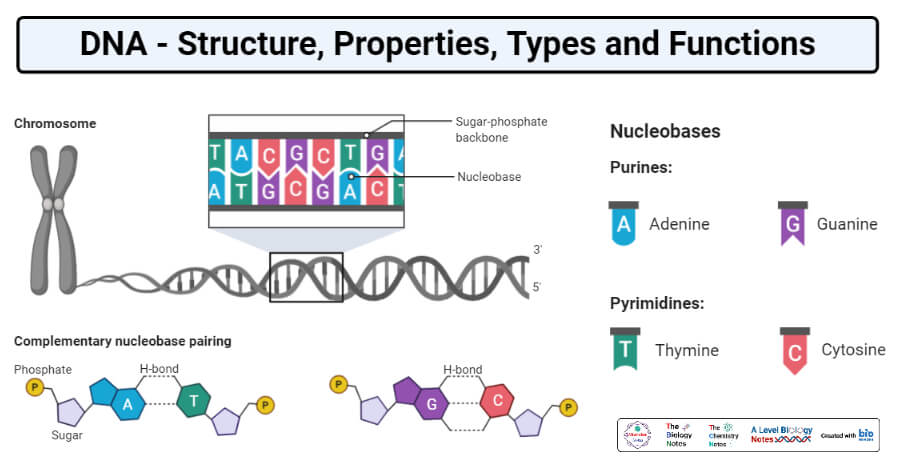
- The nucleotides present in DNA contain a 2` – deoxy – D- ribose sugar and nucleotides in RNA contain D-ribose sugar.
- The main difference is seen at the second position of the pentose structure, in the case of 2` – deoxyribose there is an absence of an alcohol group/ oxy group/ -OH group at the second position, hence the name.
- In the case of D – ribose pentose the –OH group is present at the second position.
- In both types, the pentoses are present in their β-furanose form which is a close five-membered ring structure.
Characteristics of Nucleotide
- The nitrogenous base is linked covalently to pentose sugar by an N- glycosidic bond (N-1 in the case of pyrimidines and N-9 in the case of purines is linked to the C-1 carbon atom of the pentoses.)
- The major purine bases present in DNA and RNA are Adenosine (A) and Guanine (G). The major pyrimidine bases in DNA are Thymine (T) and Cytosine (C), in RNA Thymine is replaced by Uracil (U) as a major pyrimidine base.
- The consecutive nucleotides in DNA and RNA are linked by phospho-diester linkages, the 5` phosphate group of one nucleotide is linked with the 3` hydroxyl group of another nucleotide and the complementary nucleotides are linked by hydrogen bonds.
- Adenosine and Thymine are bonded by two hydrogen bonds and Guanine and Cytosine are bonded by three hydrogen bonds.
- The backbone of the DNA is composed of the chain of nucleotides linked to each other.
Biosynthesis of Nucleotides
The nucleotides are synthesized by two pathways:
- De novo synthesis
- Salvage pathway
A. De novo synthesis of nucleotides
In this pathway, complex nucleotides are synthesized using simple molecules such as sugars and amino acids. The pathway for synthesis of Purines and Pyrimidines is different and requires different precursors.
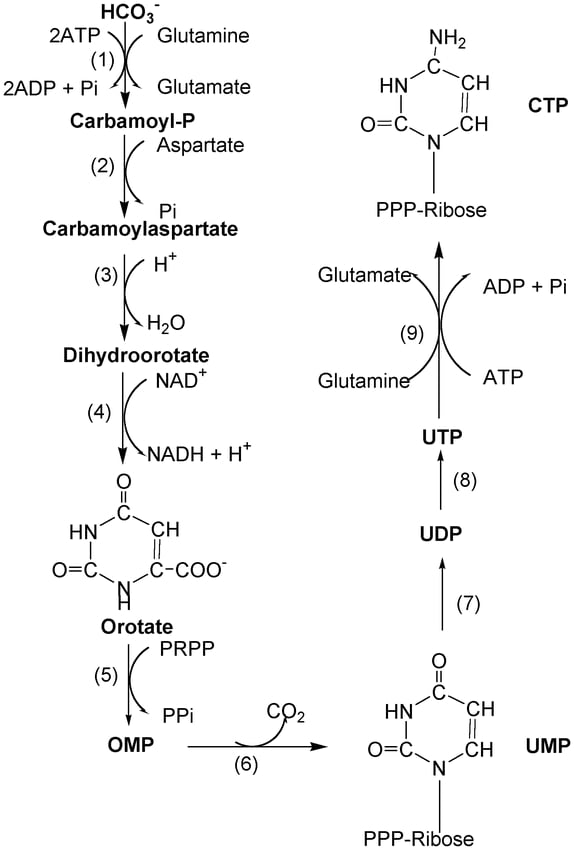
Synthesis of Pyrimidine nucleotides
The synthesis involves two major steps
- Formation of a pyrimidine ring which is a 6 membered ring structure
- Attachment of the pentose sugar (ribose – 5 – phosphate) to the ring structure
- Formation of the pyrimidine ring involves Aspartate, PRPP (5-phosphoribosyl-1-pyrophosphate) and, Carbamoyl Phosphate.
- Uracil and Cytosine are the nucleotides formed in pyrimidine synthesis. These nucleotides are formed in different stages.
- The enzymes involved in this process are:
- Aspartate transcarbamoylase (ATCase)
- Carbamoyl phosphate synthetase (Type II)
- Dihydroorotate dehydrogenase
- Orotate phosphoribosyltransferase
- Orotidine -5-phosphate carboxylase
- Glutamine, Aspartate, ATP and, CO2 are required for the synthesis of pyrimidines.
- Carbamoyl phosphate required in the biosynthesis of pyrimidine is formed in the cytosol by the enzyme carbamoyl phosphate synthetase II.
- The first step of pyrimidine synthesis is the reaction between carbamoyl and aspartate to form N-carbamoyl aspartate. This reaction is catalyzed by the enzyme Aspartate transcarbamoylase.
- One H2O molecule is removed from the N-carbamoyl aspartate to form l-dihydroorotate and this reaction is carried out by the dihydroorotase enzyme.
- This L-dihydroorotate in presence of dihydroorotate dehydrogenase forms orotate. Orotate is the most important compound to which the ribose-5-phosphate sugar is added further.
- Orotate in presence of enzyme orotate phosphoribosyltransferase forms Orotidylate. In this step the sugar, ribose-5-phosphate is provided by Phosphoribosyl pyrophosphate (PRPP). The sugar attaches to the orotate to form orotidylate.
- This Orotidylate further undergoes decarboxylation in presence of orotidylate decarboxylase to form Uridylate (UMP) also known as Uridine 5`-monophosphate.
- This uridylate is phosphorylated in further steps to form Uridine 5`- triphosphate (UTP).
- Cytidine 5`- triphosphate (CTP) is formed by the enzyme cytidine synthetase from UTP. In this conversion the nitrogen donor is glutamine.
- In this way, the biosynthesis of pyrimidines takes place.
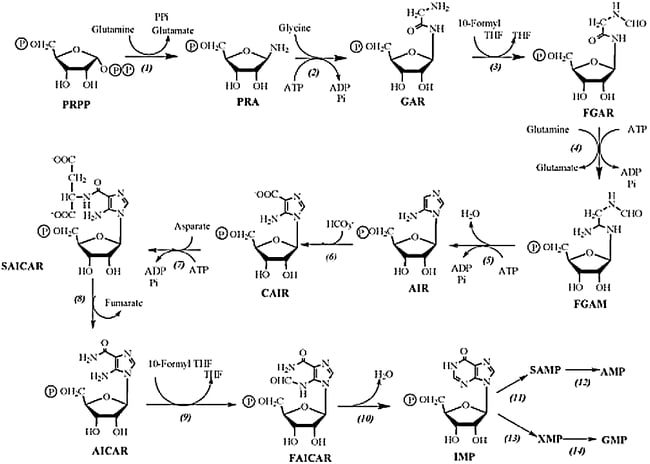
Synthesis of Purine nucleotides
- Unlike in biosynthesis of pyrimidine nucleotides, purine synthesis initiates with PRPP and then the purine ring is built around the sugar.
- The enzymes involved in this process are:
- Glutamine phosphoribosylpyrophosphate amidotransferase (Glutamine-PRPP)
- Glycinamide ribonucleotide synthase (GAR synthase)
- Gylcinamide ribotide transforamylase (GAR transformylase)
- Formylglycinamide synthase
- Aminoimidazole ribotide synthase
- Aminoimidazole ribotide carboxylase
- Succinylaminoimidazolecarboxamide ribotide synthase
- Adenylosuccinate lyase
- Aminoimidazole carboxamide ribotide transformylase
- IMP cyclohydrolase
- The first process involves the formation of Inosinate (IMP) ad further from the IMP ring the purine monophosphates are synthesized.
1. Phosphoribosyl pyrophosphate (PRPP) in presence of enzyme Glutamine-PRPP forms Phosphoribosyl amine (PRA). The amino group donated by Glutamine is attached to the C1 of the PRPP complex.
2. GAR synthase acts upon PRA to form glycinamide ribonucleotide (GAR). Here three atoms from the glycine molecule are added to the nitrogen atom.
3. GAR forms Formylglyacinamide ribonucleotide (FGAR) in presence of the enzyme GAR transformylase. The added glycine molecule is formylated by N10 – Formyl tetrahydrofolate.
4. FGAR forms formyl glycinamidine (FGAM) in presence of the enzyme FGAM synthase.
5. FGAM is catalyzed by enzyme AIM synthase to form Aminoimidazole ribonucleotide, which is further converted to Carboxyaminoimidalzole ribonucleotide (CAIR) by enzyme AIR carboxylase.
6. CAIR in presence of SAICAR synthase forms complex succinylaminoimidazole ribonucleotide. Here Aspartate donates its amino group.
7. SAICAR is converted to aminoimidazole carboxamide ribonucleotide (AICAR) in presence of the enzyme Adenylosuccinate lyase.
8. AICAR is converted to FAICAR in presence of enzyme AICAR transfromylase. The final carbon for the second ring of purine is contributed by N10 – formyl tetrahydrofolate.
9. Finally the second ring is closed to form the complete purine ring in presence of enzyme IMP cyclohydrolase to form Inosinate (IMP).
10. Now from the IMP AMP and GMP are synthesized using enzymes Adenylosuccinate synthetase and Adenylosuccinate lyase (for AMP) and IMP dehydrogenase and GMP synthetase (for GMP).
This completes the de novo synthesis of Pyrimidines.
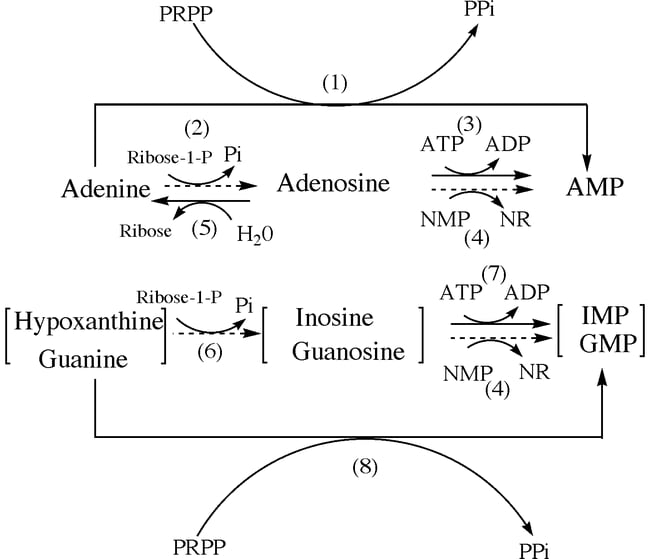
B. Salvage pathway of Purine nucleotides
- Adenine is catalyzed by the enzyme adenosine phosphoribosyltransferase (APRT) from purine bases ad purine nucleosides.
- Guanine and hypoxanthine are salvaged by the enzyme Hypoxanthine-guanine phosphoribosyltransferase (HGPRT) through the process of phosphoribosylation.
- Through other processes such as deamination, adenosine deaminase can be converted to adenosine which can further be converted to hypoxanthine.
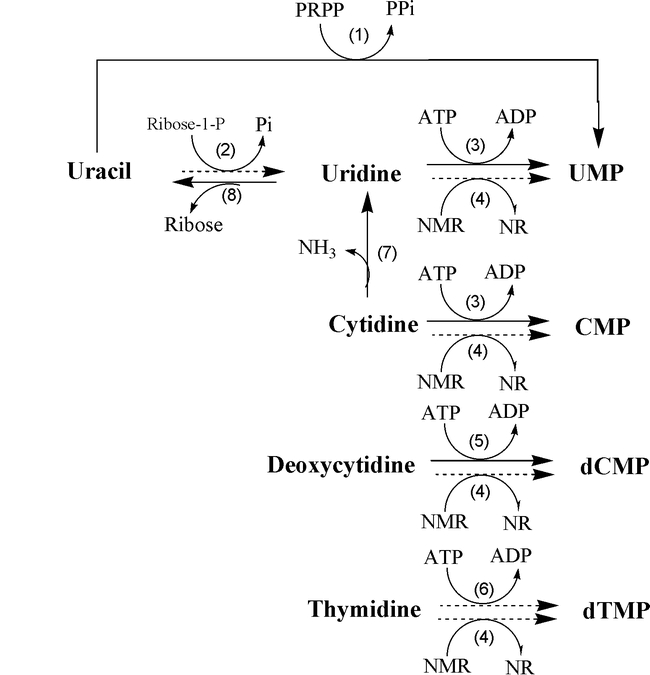
Salvage pathway of Pyrimidine nucleotides (Thymine)
- The enzyme thymidine kinase can salvage the dTTP (deoxythymidine triphosphate) from thymidine or deoxyuridine.
Pyrimidine biosynthesis regulation
- The Biosynthesis of pyrimidines is regulated at different levels. This is majorly controlled by the enzyme ATCase (Aspartate transcarbamoylase).
- ATCase is a multi-subunit allosteric enzyme with 6 active sites. It plays an important role in biosynthesis of pyrimidines. ATP acts as an activator and CTP acts as an inhibitor of ATCase.
- This enzyme brings about the conversion of aspartate to carbamoyl aspartate. Hence, when the biosynthesis of pyrimidines is to be regulated CTP binds to ATCase to stop the synthesis. This is known as feedback inhibition, where the last product of the pathway inhibits the first enzyme to regulate the synthesis. This is achieved when high levels of CTP are present in the cell.
- Biosynthesis of pyrimidines can also be regulated at PRPP (Phosphoribosyl pyrophosphate) formation. High levels of PRPP indicate high levels of pyrimidine synthesis. Thus CTP synthetase enzyme, which converts UTP to CTP is feedback inhibited by CTP. This enzyme can be activated by GTP.
- These mechanisms regulated the biosynthesis of pyrimidines.
Purine biosynthesis regulation
- The biosynthesis of purines is regulated at 3 different levels. PRPP synthetase is the first regulatory point in the pathway. Feedback inhibition is exerted by AMP and GMP over PRPP synthetase.
- PRPP amidotransferase is an allosteric enzyme and GTP, ATP, GDP, ADP, GMP and, AMP bind to the enzyme allosterically and regulates the biosynthesis.
- The last regulatory point is the branching point of IMP. Excess amount of ATP accelerated synthesis of GMP and vice versa.
Functions of Nucleotide
- Nucleotides act as energy reserves in the body and are involved n many important processes.
- ATP (Adenosine triphosphate) is considered the currency of cell and plays a crucial role in various pathways and acts as a phosphoryl group donor.
- Hydrolysis of ATP yields a large amount of energy which can be utilized by the cell of different functions.
- Nucleotides are a part of many co-factors for many enzymes.
Examples – Acetyl CoA, NAD+ (Nicotinamide adenine dinucleotide), FAD (Flavin adenine dinucleotide)
- cAMP (adenosine cyclic monophosphate) is a common second messenger produced in response to hormones and various chemical signals. cAMP is formed during a reaction of ATP catalyzed by adenylyl cyclase.
- cGMP (guanosine cyclic monophosphate) also has many regulatory functions in the cells.
- ppGpp (guanosine tetraphosphate) function is observed in bacterial cells under amino acids starvation conditions, which is in response to the slowdown of protein production.
References
- Moffatt BA, Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book. 2002;1:e0018. doi:10.1199/tab.0018.
- Zimmermann H. (2016). Extracellular ATP and other nucleotides-ubiquitous triggers of intercellular messenger release. Purinergic signalling, 12(1), 25–57. https://doi.org/10.1007/s11302-015-9483-2
- Lehninger Principles of Biochemistry – 6th ed- c2013
- Cell Biology, T Devasena, Oxford University Press.
- https://www.genome.gov/genetics-glossary/Nucleotide
- https://uh.edu/dtu/19-Nuc%20Mata-1-08.htm
- http://www2.csudh.edu/nsturm/CHEMXL153/NucleotidesCompandStruc.htm
- https://knowgenetics.org/nucleotides-and-bases/

thank you so much for simplifying
Thank you for your work Nidhi,
Ngā mihi
Rachel