DNA Library or Gene Library is simply the collection of DNA fragments cloned into vectors and stored within host organisms. They contain either the entire genome of a particular organism or the genes that are expressed at a given time.
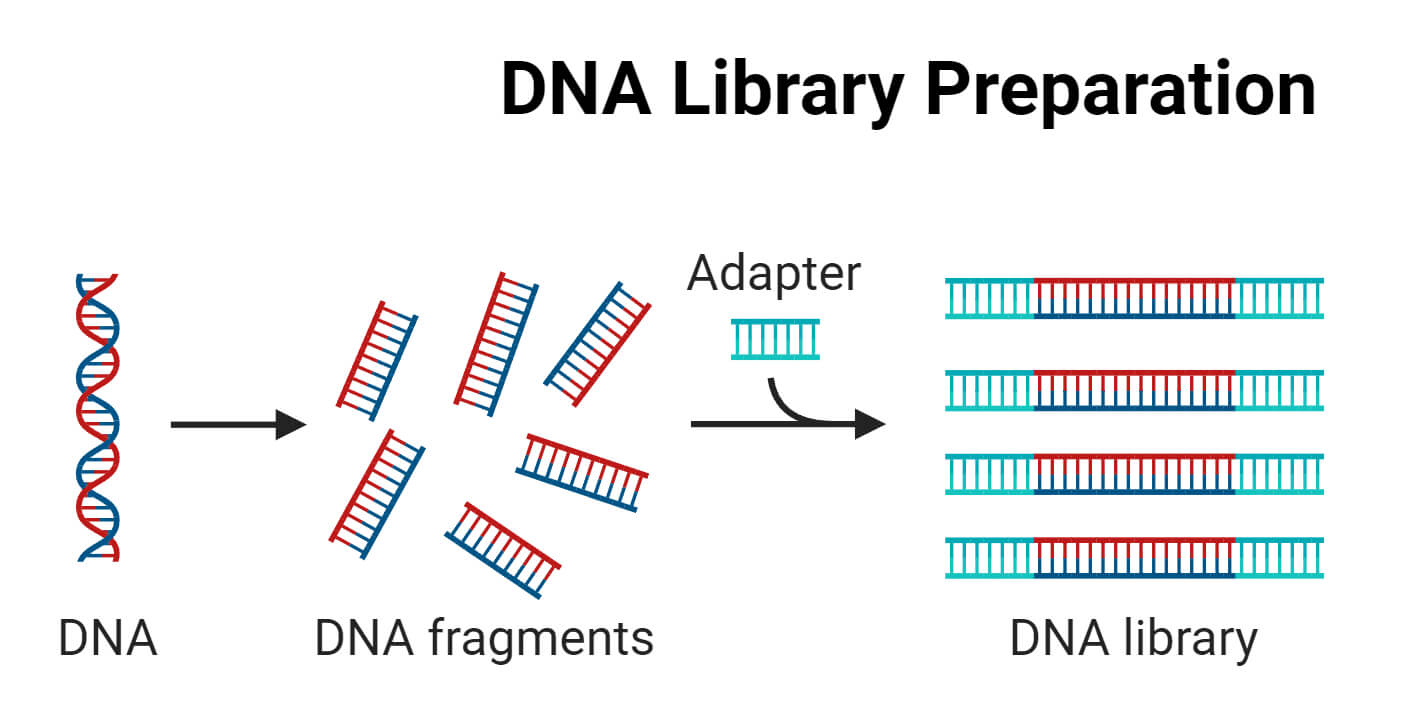
The genome is vast and complex. To understand the entire genome or study specific genes, it is important to study it in smaller and more manageable fragments. DNA libraries make the entire genome accessible in small fragments. These libraries are used to identify, isolate, and study particular genes of interest.
The development of molecular biology technologies including recombinant DNA (rDNA) technology, cloning vectors, and techniques for transforming bacteria laid the foundation for the construction of DNA libraries.
Types of DNA Library
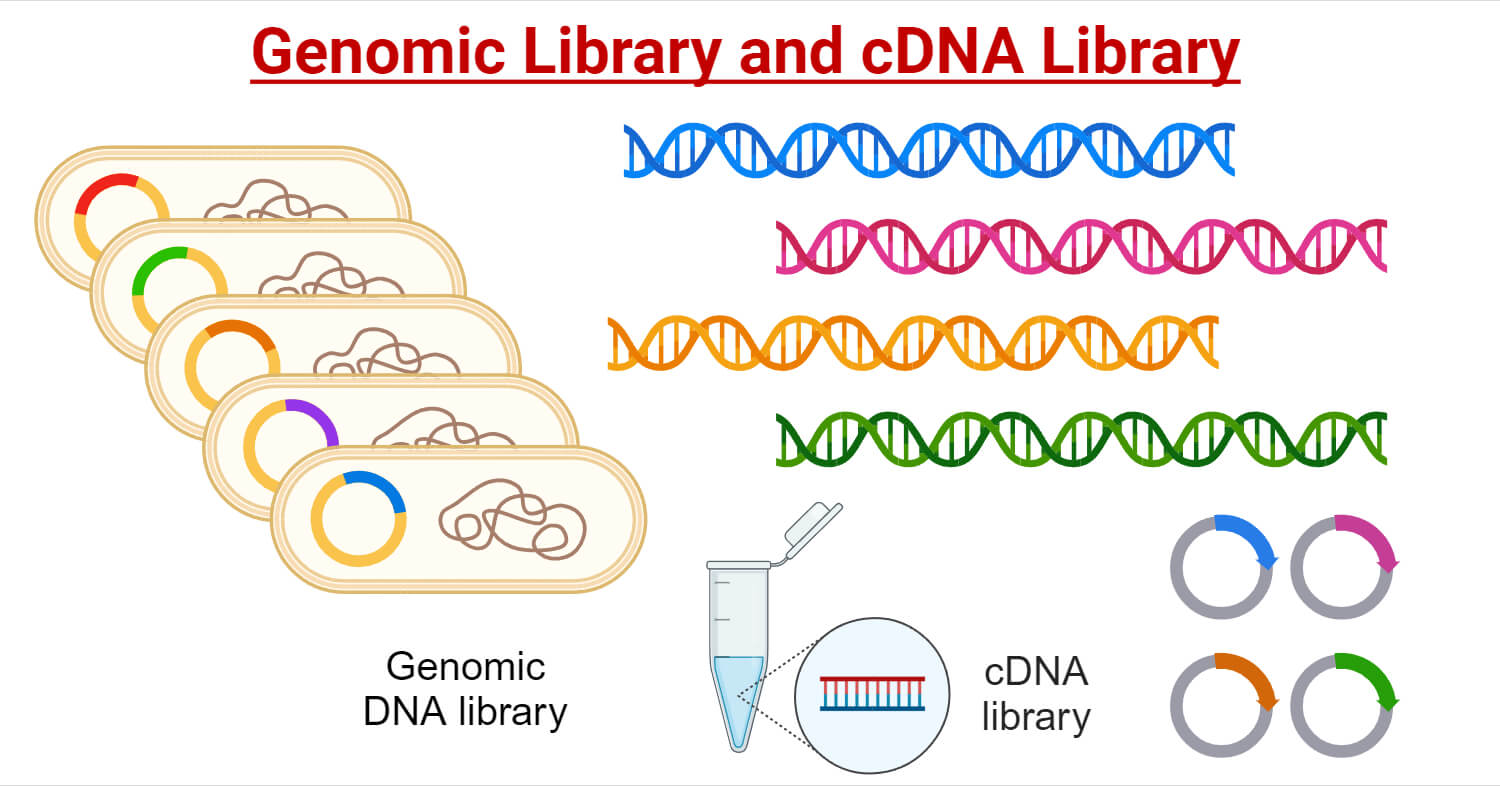
Genomic information can be obtained by two primary methods. Based on this, DNA libraries are divided into two types: genomic and cDNA library.
1. Genomic Library
- A genomic DNA library is a collection of DNA fragments that represent all genetic information of an organism. This includes both coding and noncoding regions of the DNA.
- Genomic libraries are suitable for a wide range of applications, including genome mapping and comparative genomics. It allows the study of regulatory elements and noncoding sequences that are important in gene expression and regulation.
- Genomic libraries also have certain disadvantages including the complexity and size of handling large DNA fragments and the resource-intensive process of creating and maintaining such libraries.
2. cDNA Library
- A cDNA (complementary DNA) library is a collection of cDNA molecules derived from mRNA.
- Unlike genomic libraries, cDNA libraries represent only the expressed genes of an organism, excluding non-expressed genomic regions such as introns and other noncoding sequences.
- cDNA libraries are useful for studying gene expression, protein functions, and producing recombinant proteins. Since cDNA libraries exclude noncoding regions, they provide a more focused view of the expressed genetic information.
- The disadvantages of the cDNA library include the limitation of studying gene regulation due to the absence of regulatory sequences, limited gene diversity, and bias toward highly expressed genes.
- cDNA libraries are specifically created from eukaryotes to study the expressed genes. Prokaryotes do not contain introns. Therefore, creating cDNA libraries for prokaryotes is generally not necessary, as their genomic DNA directly corresponds to their mRNA.
Differences between Genomic Library and cDNA Library
| Characteristics | Genomic Library | cDNA Library |
| Content | Genomic libraries contain DNA fragments that represent the entire genome of an organism. | cDNA libraries consist of only cDNA sequences that are actively expressed. |
| Size of DNA fragments | The DNA fragments are larger, ranging from thousands to millions of base pairs. | cDNA libraries contain smaller DNA fragments. |
| Coding and non-coding regions | It contains both coding and noncoding regions including introns and regulatory sequences. | It contains only coding regions or expressed genes. |
| Source of DNA | The source of DNA is the total genomic DNA extracted from cells or tissues. | The source of DNA for a cDNA library is mRNA which is converted to cDNA. |
| Vectors used | It uses vectors such as bacteriophages and bacterial artificial chromosomes (BACs), which can hold large DNA fragments. | cDNA libraries generally use plasmid vectors, which are suitable for the smaller cDNA fragments. |
| Application | Genomic libraries are used for genomic mapping, comparative genomics, and studying gene regulation. | cDNA libraries are particularly useful for studying gene expression patterns and the functions of expressed genes. |
Video on Genomic Library vs. cDNA Library
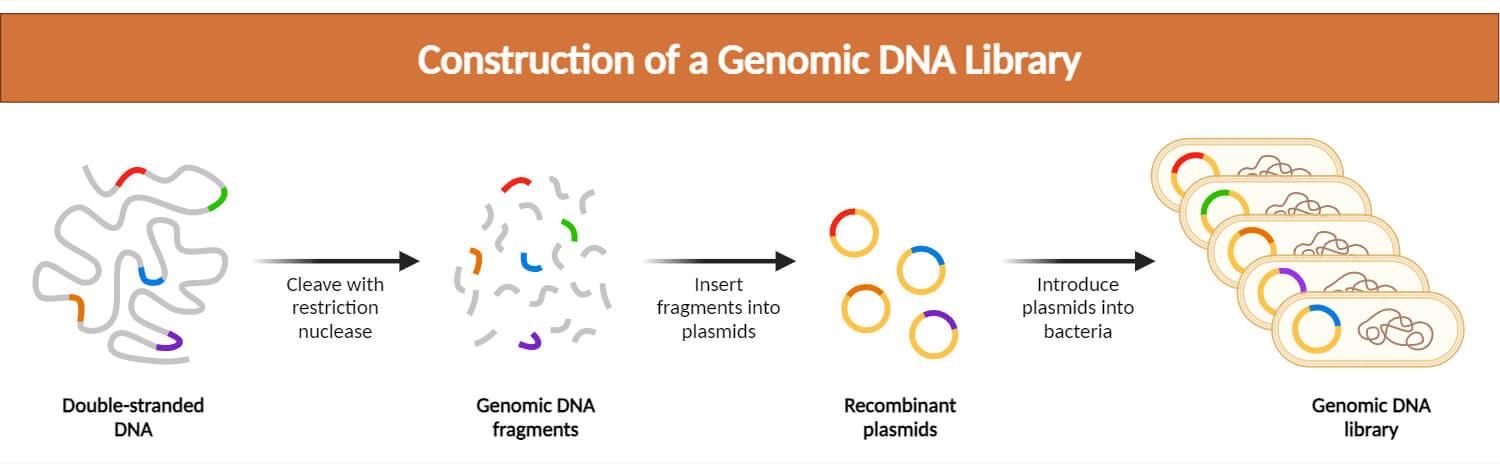
Construction (Preparation) of Genomic Library
1. Isolation of Genomic DNA
- The construction of a genomic library begins with the isolation of genomic DNA from the organism of interest.
- Genomic DNA can be isolated using different methods such as cell lysis, protein digestion, and phenol-chloroform extraction.
- The isolated DNA represents the entire genome of the organism and contains both coding and non-coding regions.
2. Fragmentation of Genomic DNA
- The isolated genomic DNA obtained is often too large to be cloned directly into vectors. So, it needs to be fragmented into smaller fragments suitable for cloning.
- This fragmentation can be achieved using physical methods such as sonication, mechanical shearing, or enzymatic methods involving restriction enzymes.
3. Cloning
- The fragmented genomic DNA is then cloned into a suitable vector. Some common vectors used for genomic library construction include plasmids, bacteriophages, bacterial artificial chromosomes (BACs), and yeast artificial chromosomes (YACs).
- Bacterial vectors are suitable for smaller DNA fragments, while YACs are used for larger DNA fragments.
- The vectors are treated with restriction enzymes to create sticky ends that are compatible with the fragmented DNA.
- DNA ligase enzyme is used to bind the DNA fragments to the vector, creating recombinant DNA molecules.
4. Transformation
- The recombinant vectors containing the cloned genomic DNA fragments are transformed into a suitable host organism, usually E. coli and yeast.
- The transformed host cells take up the recombinant vectors and are cultured on agar plates containing selective media to allow the growth of colonies containing the recombinant DNA. These colonies form a genomic DNA library.
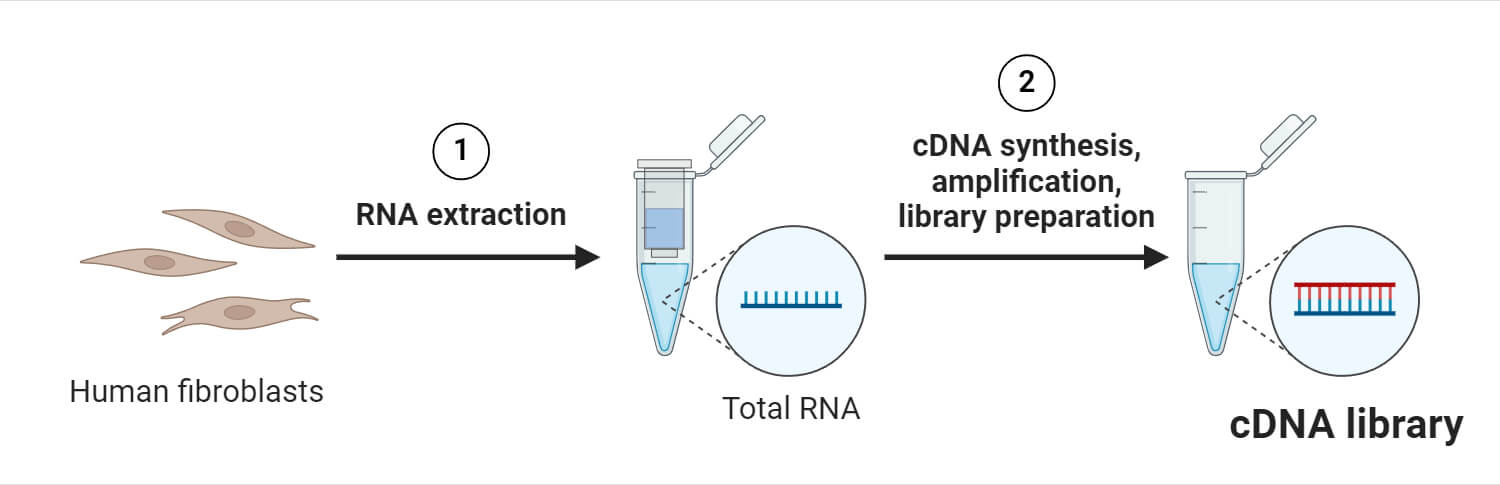
Construction (Preparation) of cDNA Library
1. Isolation of mRNA
- Construction of a cDNA library starts with the isolation of mRNA from eukaryotic cells.
- mRNA is isolated and purified using methods such as column purification. Column purification uses oligomeric dT nucleotide-coated resins that bind only mRNA with a poly-A tail.
- Eukaryotic mRNA contains a poly-A tail at the 3’ end. When a cell lysate is passed through the poly-T column, the poly-A tails of mRNA molecules bind to the oligo-dT sequences.
- This retains the mRNA in the column while all other molecules that do not have poly-A tails pass through the column and are discarded. Finally, the mRNA molecules are separated from the oligo-dT sequences using an eluting buffer.
2. Synthesis of cDNA
- After isolating and purifying mRNA molecules, the next step is the synthesis of cDNA molecules from the isolated mRNA.
- At first, a short oligo-dT primer is annealed to the 3’ poly-A tails of the mRNA molecule, which initiates the synthesis of the first DNA. Using reverse transcriptase enzyme, the primer is extended to form an RNA-DNA duplex.
- RNase H enzyme degrades the mRNA strand in the mRNA-DNA hybrid, leaving small RNA fragments that are used as primers for the synthesis of the second DNA strand.
- DNA polymerase I synthesizes the second DNA strand in segments removing the RNA primers.
- DNA ligase enzyme seals the nicks between the newly synthesized DNA fragments, resulting in the formation of a double-stranded cDNA copy of the mRNA.
3. cDNA Cloning
- The next step is the ligation of cDNA molecules into suitable vectors for cloning.
- Since cDNA has blunt ends, restriction site linkers or adapters need to be added to the ends of the cDNA molecules to make them compatible with the vector DNA. These linkers contain recognition sites for restriction enzymes. The linkers are then digested using restriction enzymes.
- The most commonly used vectors for cloning cDNA are plasmid and phage vectors. The suitable vectors are cut with the same restriction enzyme as the cDNA.
- The cDNA molecules are joined with the vector DNA which creates recombinant DNA molecules.
4. Transformation
- The recombinant DNA molecules are transformed into host cells that can be cultured to produce colonies containing the cloned cDNA inserts.
- To select host cells that have successfully taken up the recombinant DNA, the transformed cells are cultured on agar plates containing a selective medium. The selective medium contains antibiotics that inhibit the growth of untransformed cells.
- The presence of a selectable marker gene on the vector ensures that only cells containing the recombinant DNA survive and form colonies on the selective medium. The resulting colonies form the cDNA library.
Screening a DNA Library
- After the construction of gene libraries, the screening process for both cDNA and genomic libraries is important to identify and isolate the recombinant clones containing the DNA inserts of interest.
- There are several screening methods. The most commonly used screening method for gene libraries is hybridization-based screening. This method uses labeled DNA or RNA probes that are complementary to the target DNA sequence in the gene library.
- In hybridization-based screening, the colonies or plaques in the gene libraries are transferred onto a solid membrane such as a nitrocellulose membrane.
- The membrane is then probed with radiolabeled probes to identify clones containing the target sequence. The probe will hybridize with its complementary sequence in the target DNA.
- Positive clones are identified based on the presence of hybridization signals, which are visualized using autoradiography or fluorescence detection.
- Other commonly used screening methods include PCR screening, immunological assay, and sequencing-based screening.
Applications of DNA Library
Applications of genomic libraries
- Genomic libraries are essential for constructing physical maps of genomes which helps in understanding the layout and structure of genes.
- These libraries are particularly useful for studying non-coding regions, regulatory elements, and gene sequences that may not be expressed.
- Genomic libraries from different species can be compared to study their evolutionary relationships.
- Genomic libraries also help identify genetic mutations and disease-associated genes which is important for understanding the genetic basis of diseases.
Applications of cDNA libraries
- cDNA libraries are useful for studying actively expressed genes in different tissues under specific conditions. cDNA library allows the identification and cloning of expressed genes.
- cDNA libraries can be used to compare gene expression profiles between different species which is useful in the study of evolutionary biology.
- cDNA libraries are also used to produce recombinant proteins.
- These libraries are also used to identify and study the expression of disease-related genes, which can help in the development of diagnostic markers.
References
- Libretexts. (2021, March 6). 3.6: CDNA and Genomic libraries. Retrieved from https://bio.libretexts.org/Bookshelves/Biochemistry/Supplemental_Modules_(Biochemistry)/3._Biotechnology_1/3.6%3A_cDNA_and_Genomic_Libraries
- Libretexts. (2021, March 6). 3.6: CDNA and Genomic libraries. Retrieved from https://bio.libretexts.org/Bookshelves/Biochemistry/Supplemental_Modules_(Biochemistry)/3._Biotechnology_1/3.6%3A_cDNA_and_Genomic_Libraries
- Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26837/
- Dale, J., Plant, N., & Schantz, M. V. (2013). From Genes to Genomes: Concepts and Applications of DNA Technology. Wiley.
- Brown, T. A. (2010). Gene cloning and DNA analysis: An introduction (6th ed.). Wiley-Blackwell.
- Chauhan, T. (2022, February 2). Genomic DNA Library- Preparation and applications. Retrieved from https://geneticeducation.co.in/genomic-dna-library-preparation-and-applications/
- Raj, A. (2016, November 2). Genomic and CDNA Libraries | Genetics. Retrieved from https://www.biologydiscussion.com/genetics/genomic-and-cdna-libraries-genetics/61424
- Exploring screening methods in recombinant DNA technology | Danaher Life Sciences. (2024, June 3). Retrieved from https://lifesciences.danaher.com/us/en/library/recombinant-dna-screening.html
- DNA Libraries | Thermo Fisher Scientific – DE. (n.d.). Retrieved from https://www.thermofisher.com/de/de/home/life-science/cloning/cloning-applications/library-construction.html

Thanks for sharing
thanks great work
Nice website