In molecular biology, a vector refers to a DNA molecule that transfers genetic material into another cell and the genetic material gets to incorporate in the cell genome and expressed. The foreign DNA carried by the vector is called recombinant DNA. The vectors are generally characterized by three factors such as the origin of replication, multi cloning site, and a selective marker. Plasmids and bacteriophage are commonly used molecular vectors which are described as following.
Interesting Science Videos
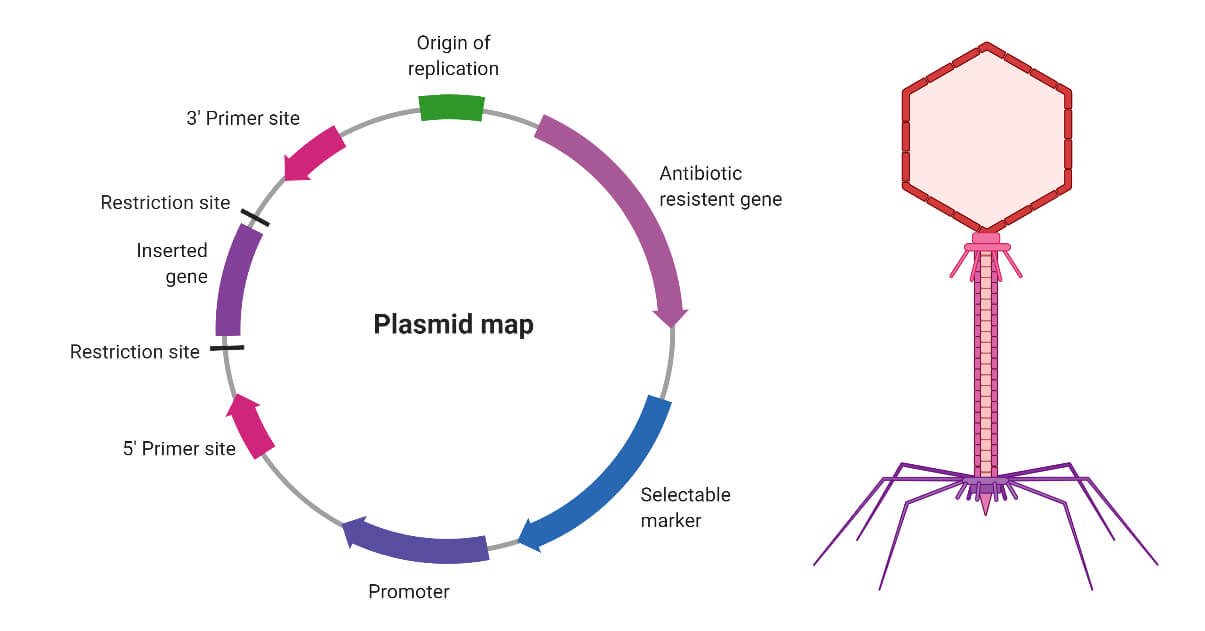
Plasmids
Plasmids are circular DNA molecule which are present independently inside the bacterial cell and has one or more genes. These genes often play an important role in the host bacterium. To illustrate, bacterial plasmids mostly contain antibiotic-resistant genes which protects the bacteria from antibiotics like ampicillin. In genetic engineering antibiotic-resistant gene functions as a selective marker, to find out whether the plasmid is transferred in the bacterial cell. Apart from that, most of the plasmids contain a specific DNA sequence called the origin of replication which helps plasmids to replicate independently. Some plasmids often used host cells enzyme for replication while some can code for their own special type of enzymes for replication.
Factors important for selecting a vector
Size and copy number
The size of the plasmids ranges from 1kb to 250 kb. But the size of the plasmid is important for cloning purposes. All types of plasmids are not useful as a cloning vector; plasmids with a size of fewer than 10 kb can be used as vectors. However, in some cases, larger plasmids can also involve as vectors. Copy number refers to the number of plasmid molecules present in a particular bacterial cell. Larger plasmids are generally considered as stringent and often present in low numbers while small plasmids are generally present in high numbers. The high number of plasmids helps to incorporate several recombinant DNA into a cell.
Conjugation and Compatibility
Conjugation is a process by which bacterial plasmid can be transferred from one cell to another cell. Conjugative plasmids involve in the process of sexual conjugation. On the other hand, non-conjugative plasmids are also present in bacterial cells. The non-conjugative plasmids lack a type of gene called tra gene which is involved in the transfer of plasmids by conjugation and present in conjugative plasmids. A bacterial cell can have several types of plasmids among which different types of conjugative plasmids are also present. In this case, the transfer process required the compatibility of the plasmids present in a bacterial cell. Two incompatible plasmids will not be present inside a cell; one or the other will be lost. Hence, the compatibility of the plasmids plays an important role in the selection of cloning vectors.
Types of Plasmids
The naturally occurring plasmids are classified under five major groups on the basis of the genes present in the plasmids. The five groups are the following:
F plasmid or Fertility: F plasmid contains specifically the tra gene and facilitates the transfer of plasmids by conjugation.
R plasmid or Resistance: This type of plasmids contains one of the multiple antibiotic resistance genes. In clinical microbiology, R plasmid plays an important role as it is the main reason behind the development of antibiotic resistance in bacterial pathogens. An example of R plasmid is RP4 which is commonly present in Pseudomonas sp.
Col Plasmid: Col plasmids are responsible for the production of colicin and commonly found in some strains of E. coli. This is a type of toxin which is generally released in the environment to reduce competition for nutrition. ColE1 is an example of this type of plasmid which is found in E. coli.
Degradative plasmid: This type of plasmid is responsible for the degradation of complex compounds like toluene, salicylic acid. Example: TOL found in Pseudomonas putida.
Virulence plasmid: Virulence plasmids are present in pathogenic bacteria and responsible for causing infection or disease to the host cell. Ti plasmid is an example of virulence plasmid which is present in Agrobacterium tumefaciens and causes crown gall disease in dicotyledonous plants.
Bacteriophage
Bacteriophages are a group of viruses that used bacterial cells as host and reproduce by infecting bacterial cells. Like all other viruses bacteriophages are also consisting of a protein coat that provides protection to the genome. Most of the phages are DNA viruses that code for several genes responsible for virus replication. A major problem of using plasmids as a cloning vector is the size of the gene of interest is small. However, bacteriophages help to overcome this problem, as larger genes can be transferred by using bacteriophage as a cloning vector.
Lytic and Lysogenic Phages
The pattern of infection of different bacteriophages is mostly needed three steps to complete. Firstly, the attachment of the phage at a specific site of bacterial takes place. The phage is then injected into the DNA inside the bacterial cell. In the second step, the genes present in the DNA of the phage codes several enzymes which helps to replicate the phage DNA. Finally, other genes also code several proteins by which the capsid is formed and the new phage particle is assembled inside the capsid. This step is followed by lysis of the cell and release of the phages from the cell. In several bacteriophages, this cycle occurs rapidly and it is called a lytic cycle. On the other hand, in some phages, the phage DNA got incorporated in the host genome after entering the host cell and remains there for several cell divisions. The phage DNA which has incorporated in the bacterial DNA is called prophage and it is quiescent. The bacteria which carry the phage DNA remain as same as any other uninfected cells. This type of infection is called a lysogenic cycle. The phage DNA finally released from the host cell by entering into the lytic cycle. The infection cycle of lambda phage is an excellent example of a lysogenic cycle. In the case of cloning, the lysogenic cycle is most important, because, the phage DNA can carry a specific gene that is further incorporated in the host genome. As a result, the host genome will code a specific gene carries by the phage. Therefore, lambda phages are mostly used as a cloning vector.
Genetic organization of lambda DNA
The lambda phage is a typical example of bacteriophage which has a polyhedral head and tail. The DNA is protected by the polyhedral structure, and the tail interacts with the specific site of bacteria for attachment. Lambda phage usually infects E. coli. The DNA of lambda phage is 49 kb of size. Gene mapping and DNA sequencing analysis provide all the information regarding the genetic arrangement of the phage DNA. According to the data gathered from these analyses, the genes of similar functions are clustered together in the genome. To illustrate, the genes responsible for the formation of the capsid are clustered together at the left-hand side of the polyhedral head while the genes responsible for the incorporation of the phage DNA with the host genome clustered together at the middle.
Linear and circular forms of lambda DNA
The formation of the lambda DNA plays an important role as a cloning vector. The genome of a lambda phage is made up of two complementary linear DNA. As the DNA strands are complementary they attach together and forms circular double-stranded DNA. In molecular biology, the term sticky ends refer to the complementary single strands as they attach together. In lambda DNA the sticky end is termed as cos site which plays essential roles in the infection cycle. Firstly, the phage DNA remains circular inside of a bacterial cell after insertion due to the cos sites. Then, the second role of the cos sites is rather different and comes into play after the prophage has excised from the host genome. At this stage, a large number of new lambda DNA molecules are produced by the rolling circle mechanism of replication, in which a continuous DNA strand is “rolled off” the template molecule. The result is a catenane consisting of a series of linear lambda genomes joined together at the cos sites. The role of the cos sites is now to act as recognition sequences for an endonuclease that cleaves the catenane at the cos sites, producing individual lambda genomes. This endonuclease, which is the product of gene A on the lambda DNA molecule, creates the single-stranded sticky ends and also acts in conjunction with other proteins to package each lambda genome into a phage head structure. The cleavage and packaging processes recognize just the cos sites and the DNA sequences to either side of them, so changing the structure of the internal regions of the lambda genome, for example by inserting new genes, has no effect on these events so long as the overall length of the lambda genome is not altered too greatly.
M-13: Filamentous phage
M-13 is a type of bacteriophage and it is totally different from the lambda phages. The structure is filamentous, the genome size is very small (6407 nucleotides), single-stranded and remain in a circular formation. Due to the small size of DNA, the phage genome can carry only a few recombinant genes. The M-13 phage DNA enters inside an E. coli cell via pilus. Inside the cell, the phage DNA codes for specific enzymes that synthesize a complementary strand of DNA. This newly formed double-stranded DNA molecule then proliferates into several copies. During the bacterial cell division, each daughter cell contains copies of phage DNA and the phage DNA continues to replicate inside the daughter cells. Finally, new phage particles are assembled and released from the bacterial cell. M-13 phages are also used as a cloning vector. The genome size is 10 kb which is enough to insert new genes. Apart from that, the modification of single-strand DNA to double-strand DNA inside the host makes it unique.
References
- T.A. Brown, Gene cloning and DNA analysis: An Introduction, 6th Edition, Wiley-Blackwell.
- Bolivar, F. and Backman, K., 1979. [16] Plasmids of Escherichia coli as cloning vectors. In Methods in enzymology (Vol. 68, pp. 245-267). Academic Press.
- Meissner, P.S., Sisk, W.P. and Berman, M.L., 1987. Bacteriophage lambda cloning system for the construction of directional cDNA libraries. Proceedings of the National Academy of Sciences, 84(12), pp.4171-4175.
Internet Sources
- 3% – https://www.slideshare.net/PradeepNarwat/vectors-93868589
- 2% – https://www.slideshare.net/Graanwatan/gene-cloning-by-ta-brown-6th-ed
- 1% – https://encyclopedia2.thefreedictionary.com/Plasmids
- 1% – https://en.wikipedia.org/wiki/Episome
- 1% – https://en.m.wikipedia.org/wiki/Colicin
- <1% – https://www.sciencedirect.com/science/article/pii/S0734975015000634
- <1% – https://www.sciencedirect.com/science/article/pii/B9780123946263000028
- <1% – https://www.ocf.berkeley.edu/~pahc/Tests/MCB/MCB102/Phillip/Su09PS6KEY.pdf
- <1% – https://www.fda.gov/media/108025/download
- <1% – https://www.chegg.com/homework-help/definitions/cloning-vectors-14
- <1% – https://quizlet.com/52970793/chapter-9-bacterial-genetic-analysis-flash-cards/
- <1% – https://medical-dictionary.thefreedictionary.com/nonconjugative+plasmid
- <1% – https://en.wikipedia.org/wiki/Plasmid
- <1% – https://en.wikipedia.org/wiki/Episomes
- <1% – https://en.wikibooks.org/wiki/Structural_Biochemistry/DNA_recombinant_techniques/History_and_Study_of_Bacteriophage_Lambda
- <1% – https://courses.lumenlearning.com/sanjacinto-biology1/chapter/dna-replication-prokaryotes/
- <1% – https://courses.lumenlearning.com/boundless-microbiology/chapter/cloning-techniques/
- <1% – https://answers.yahoo.com/question/index?qid=20081129161554AAQunVw
- <1% – http://www.web-books.com/MoBio/Free/Ch1F1.htm
- <1% – http://www.shareyouressays.com/knowledge/biology-question-bank-131-mcqs-on-genes-chromosomes-answered/114627
- <1% – http://europepmc.org/articles/PMC4270446
