DNA cloning is a method used to produce multiple identical copies of a DNA fragment within a cell. DNA cloning is also known as gene cloning or molecular cloning. All three terms are used interchangeably to describe the single technique.
DNA cloning has opened up new possibilities in areas like genetic engineering and biomedical research. With DNA cloning, researchers can study gene function and explore different biological processes.
Interesting Science Videos
Principle of DNA Cloning
The principle of DNA cloning involves the production of multiple copies of a particular DNA fragment of interest. It involves inserting the desired DNA fragment into a cloning vector, usually a plasmid, to create a recombinant DNA molecule which is then introduced into host cells through transformation. The transformed cells are selected and cultured on selective media, allowing for the replication of the inserted DNA fragment. This results in the production of multiple copies of the desired DNA, which can be isolated for further analysis.
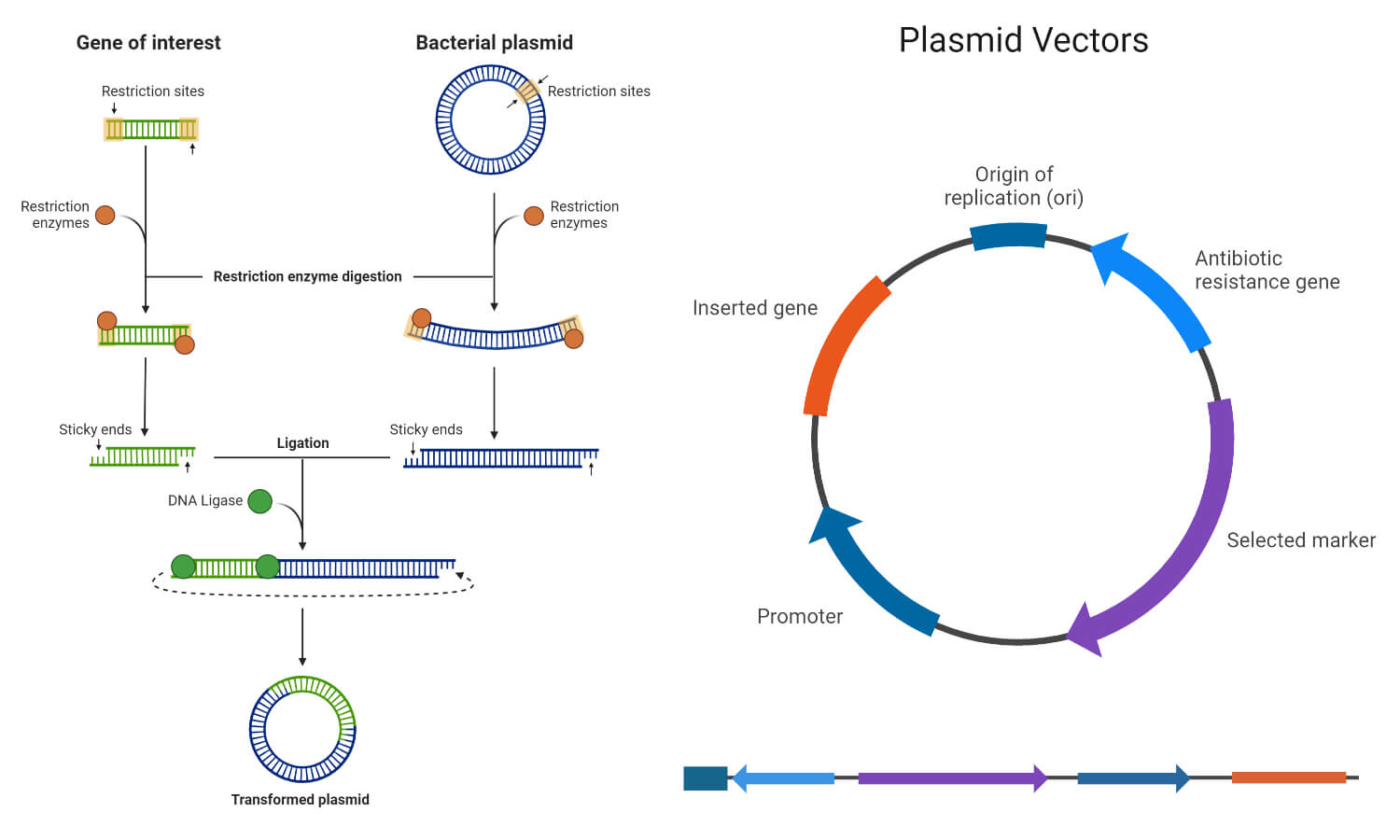
Steps in DNA Cloning
The process of DNA cloning can be divided into the following five steps:
1. Preparation of gene of interest and vector
- The first step in DNA cloning is to obtain the gene of interest, which contains the desired DNA sequence to be cloned.
- For simple organisms, such as bacteria, the DNA fragment can be obtained by digesting the genomic DNA using restriction enzymes. More complex organisms, such as mammals, use alternative methods such as reverse transcription of mRNA or PCR amplification. In some cases, physical methods like sonication or shearing can be used to fragment DNA.
- The gene of interest is inserted into a vector and the most commonly used vector is the plasmid, a circular DNA molecule commonly found in prokaryotes.
- Both the vector and the gene of interest are cut using the same or compatible restriction enzymes. Restriction enzymes identify specific DNA sequences and cut the DNA at those sites.
2. Ligation of the gene of interest and vector
- After digestion with restriction enzymes, the vector and the gene of interest can be joined together to form recombinant DNA (rDNA) using the enzyme DNA ligase.
- DNA ligase works by recognizing and binding to the ends of the DNA fragments that are cut by the restriction enzymes. It then catalyzes the formation of new phosphodiester bonds, joining the DNA fragments.
3. Transformation
- The next step is transformation where the rDNA is introduced into a host cell that takes up and expresses the inserted gene of interest.
- Before the transformation, the host cells must be made competent, which means they are made capable of taking up DNA through their membrane.
- Different methods are used to make host cells competent. One common approach involves using cold calcium chloride, followed by a brief heat shock.
- Another method for achieving competency is through electroporation. In electroporation, the host cells are exposed to an electric field, which creates temporary pores in the cell membrane, making it more permeable to DNA molecules.
- Once the host cells have been made competent, the recombinant plasmid is mixed with the competent host cells. The transformed host cells then take up the recombinant plasmid and incorporate it into their own genetic material.
4. Selection/screening and culturing of transformed cells
- After the transformation of host cells with the recombinant plasmid, the next step is the selection or screening of the transformed cells.
- The transformed host cells are plated onto a nutrient agar medium that contains a specific antibiotic. The antibiotic is chosen based on the antibiotic resistance gene present in the recombinant plasmid.
- Successfully transformed cells will contain genes with antibiotic resistance, allowing them to grow and form colonies on the selective media.
5. Isolation of recombinant DNA
- Once colonies of transformed cells have formed on the agar plate, the rDNA from the culture can be isolated.
- A single colony of the transformed cells is selected from the agar plate and cultured in a liquid nutrient medium.
- During this process, the host cells will multiply, and the recombinant plasmid, along with its inserted gene of interest, will also be replicated, producing multiple copies of the rDNA.
- The isolated rDNA can be further analyzed and used for various applications, such as protein expression or genetic engineering experiments.
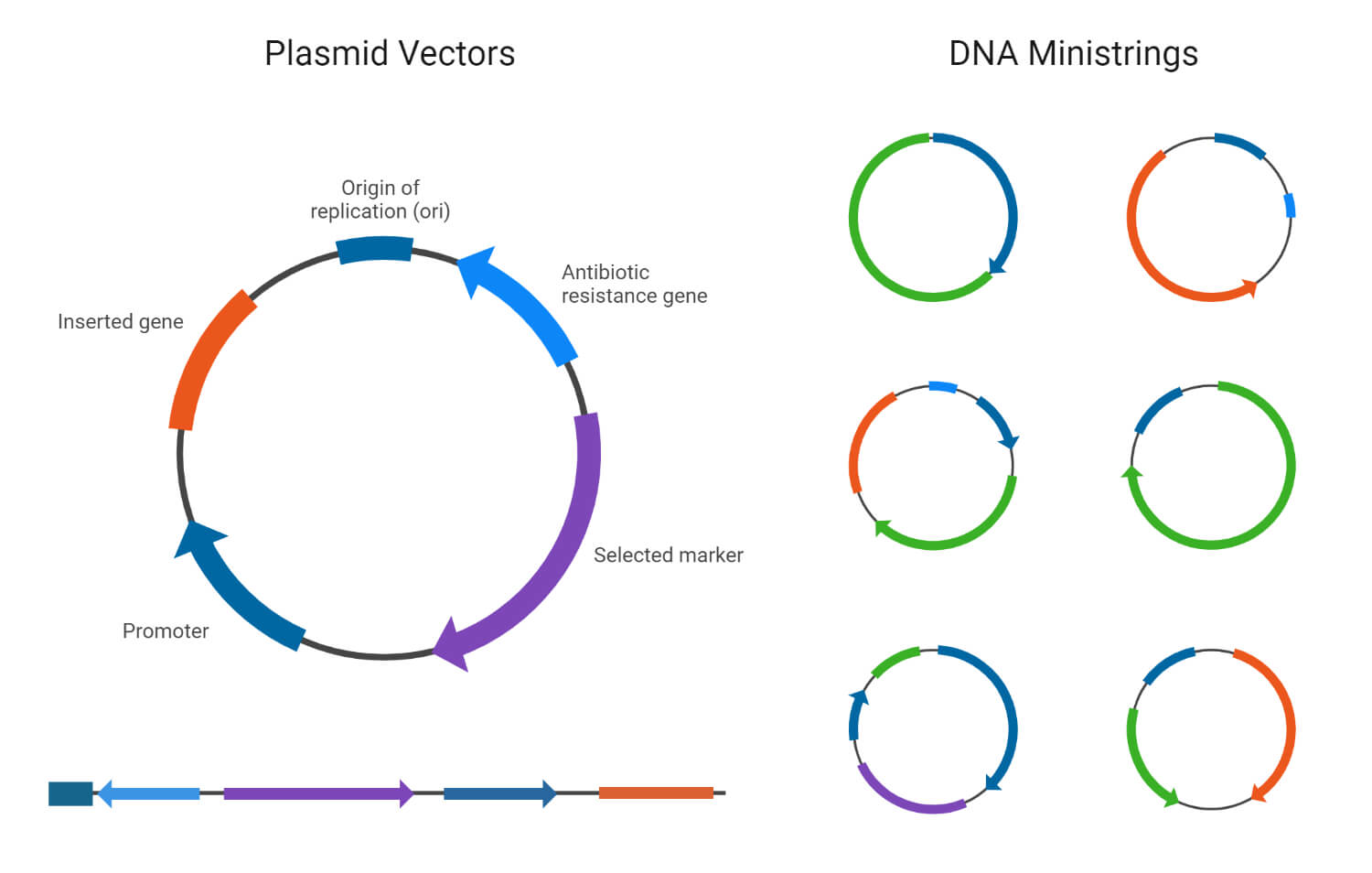
Components of DNA Cloning
1. Cloning vector
- A cloning vector is a DNA molecule used as a carrier to insert a specific segment of foreign DNA into a host cell for cloning.
- In order to be used as a cloning vector, a DNA molecule must have specific characteristics. One important feature is the ability to replicate within the host cell. It should also be small, usually less than 10 kilobases (kb), for easy handling and stability.
- A cloning vector also requires a suitable cloning site and a selectable marker that is recognized by specific restriction enzymes to insert DNA fragments into the vector.
There are different types of cloning vectors. Some commonly used cloning vectors are:
- Plasmids are circular, autonomously replicating DNA molecules widely used as cloning vectors. They can hold DNA inserts of sizes up to around 15 kb.
- Bacteriophages, also known as phages, are viruses that infect bacteria. Bacteriophages like lambda (λ) and M13 are often used as cloning vectors.
- Cosmids are hybrid vectors that combine the characteristics of both plasmids and bacteriophages. They are more stable than regular plasmids.
- Bacterial artificial chromosomes (BACs) are large cloning vectors used for cloning DNA sequences in bacterial cells and they can hold DNA segments up to 350 kb.
Yeast artificial chromosomes (YACs) are vectors used for cloning DNA fragments larger than 1 megabase (1 Mb) in size. They are commonly used in genome mapping and sequencing projects.
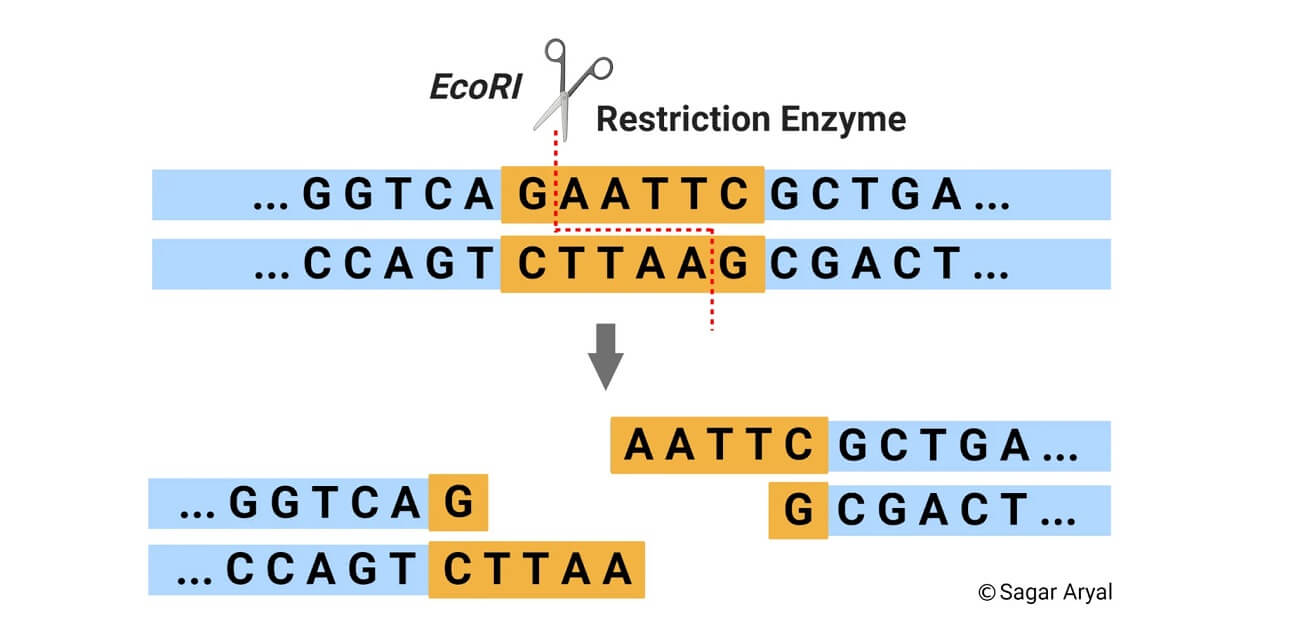
2. Restriction enzymes
- Restriction enzymes, also called restriction endonucleases, are enzymes produced by bacteria that recognize and cut DNA sequences at unique sites called recognition sites.
- Different restriction enzymes have different cutting patterns, which result in either sticky ends or blunt ends.
- Sticky ends result in overhanging single-stranded DNA sequences that can easily be joined with other DNA fragments that are cut by the same enzyme. On the other hand, blunt ends have no overhangs, requiring additional techniques or enzymes for successful ligation.
DNA cloning methods
There are several methods of DNA cloning. Some of the popular cloning methods are:
1. Traditional Cloning
- Traditional cloning, also called restriction enzyme-based cloning, uses restriction enzymes to cut the DNA insert and vector at specific restriction sites.
- The DNA insert should not contain any internal restriction sites that are similar to the ones present on the plasmid as it could result in the production of unwanted smaller DNA fragments.
- After the DNA fragments have been cut by the restriction enzymes, DNA ligase is used to join the DNA insert with the vector.
2. PCR Cloning
- PCR cloning is a type of cloning that involves the direct ligation of DNA fragments, obtained through PCR amplification, into a vector without the need for cutting the insert using restriction enzymes.
- There are several types of PCR cloning methods. One popular method of PCR cloning is TA cloning.
- In TA cloning, Taq polymerase adds an adenine (A) residue to the 3′ ends of PCR products, creating “A-tailed” DNA fragments. These fragments are directly ligated with “T-tailed” vectors having thymidine (T) residues at their ends using DNA ligase.
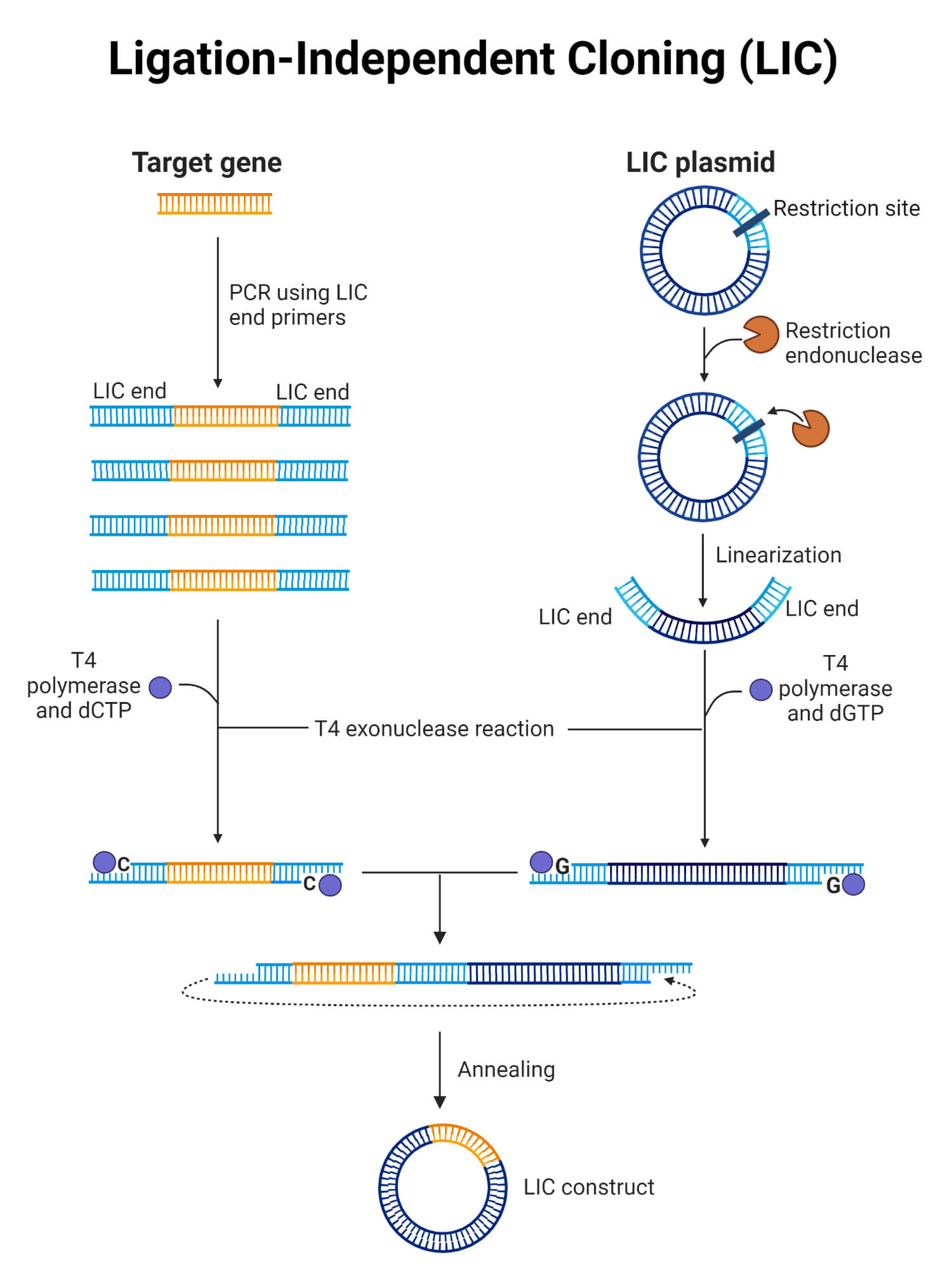
3. Ligation-Independent Cloning (LIC)
- Ligation-independent cloning (LIC) is a method where specific short sequences are added to the ends of a DNA insert to match the sequences on a vector.
- The 3′ ends of the DNA fragment are trimmed using the enzymes with 3′ to 5′ exonuclease activity which creates cohesive ends between the DNA insert and the vector.
- The vector and insert molecules are combined. The resulting plasmid contains four single-stranded DNA nicks which are repaired by the host during transformation.
- An important advantage of LIC is that it maintains the original sequence integrity without introducing any additional elements.
4. Seamless Cloning (SC)
- Seamless cloning (SC) is a method that relies on matching short sequences at the ends of a DNA fragment with corresponding short sequences on a vector. It is similar to the LIC method.
- In SC, an enzyme with 5′ to 3′ exonuclease activity is used to create 3′ overhangs on the DNA fragment.
- The advantage of SC over traditional cloning is that they enable the insertion of multiple DNA fragments into a vector.
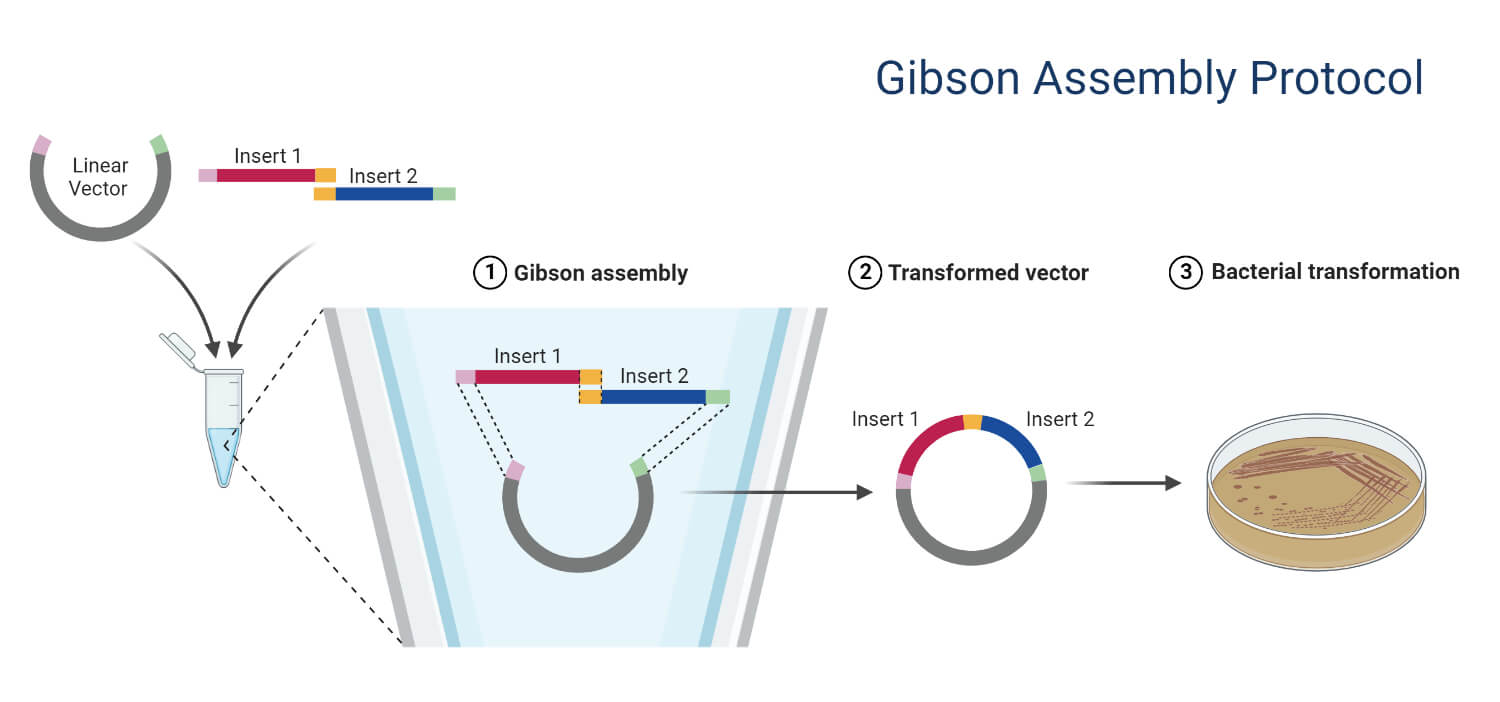
5. Recombinational Cloning
- Recombinatorial cloning involves the use of site-specific DNA recombinases that facilitate the exchange and recombination of DNA fragments at specific recombination sites.
- The process begins by inserting a DNA fragment into an entry vector, creating an entry clone. Once the entry clone is obtained, it is recombined with a destination clone.
- Recombinatorial cloning provides an efficient way to create complex DNA constructs by allowing easy transfer of DNA fragments between different vectors through site-specific recombination.
Applications of DNA Cloning
DNA cloning has many applications in various fields of research. Some major applications along with examples of DNA cloning are:
- DNA cloning is useful for studying the gene functions of specific genes in different organisms. For example, the cloning of the green fluorescent protein (GFP) gene from jellyfish has allowed the visualization of protein expression in living cells.
- DNA cloning has been used for producing recombinant proteins in large quantities. For example, the cloning of the human insulin gene led to the large-scale production of insulin for the treatment of diabetes, reducing dependence on animal-derived insulin.
- DNA cloning plays an important role in genetic engineering to create genetically modified organisms (GMOs), which introduces desired genes into organisms to modify their traits. For example, cloning genes to create genetically modified crops with improved traits such as pest resistance and higher yield.
- DNA cloning is also useful in gene therapy, where therapeutic genes are cloned and used to treat genetic diseases.
- DNA cloning techniques are also used in forensic analysis. Cloning specific DNA regions can be used in the amplification and analysis of genetic markers to determine an individual’s identity in forensic investigations.
Challenges and Limitations of DNA Cloning
DNA cloning has brought significant advancements in various fields, but it also has its limitations that need to be considered. Some of the challenges and limitations are:
- Traditional DNA cloning can be time-consuming, especially when working with large DNA fragments. It may take several days to complete steps such as culturing and restriction digestion.
- Another limitation is the potential for contamination during the cloning process.
- DNA cloning can be costly and labor-intensive due to the reagents, enzymes, and equipment required.
- In order to ensure successful cloning, the compatibility between the insert and vector needs to be considered.
Ethical Considerations in DNA Cloning
DNA cloning raises several ethical concerns. To ensure ethical practices in DNA cloning, it is important to address and take into account these ethical concerns and considerations.
- One of the concerns is genetic modification, which raises questions about the potential consequences for organisms and ecosystems.
- Introducing cloned or genetically modified organisms (GMOs) into the environment can have unintended environmental impacts that need to be carefully assessed.
- Another ethical issue is patenting and commercialization of genetic resources, which may negatively impact scientific research and access to genetic information.
- Privacy of genetic information is also an important consideration, with concerns about confidentiality and potential misuse of individuals’ genetic data. Informed consent is also crucial when human subjects are involved in cloning research.
References
- 9.3: Cloning and Recombinant Expression – Biology LibreTexts
- Amarakoon, I. I., Hamilton, C.-L., Mitchell, S. A., Tennant, P. F., & Roye, M. E. (2017). Biotechnology. Pharmacognosy, 549–563. doi:10.1016/b978-0-12-802104-0.00028-7
- Bacterial Artificial Chromosome (BAC) (genome.gov)
- Bertero, A., Brown, S., & Vallier, L. (2017). Methods of Cloning. Basic Science Methods for Clinical Researchers, 19–39. doi:10.1016/b978-0-12-803077-6.00002-3
- Brown, T. (2020). Gene Cloning and DNA Analysis (8th ed.). Wiley.
- Carter, M., & Shieh, J. C. (2010). Molecular Cloning and Recombinant DNA Technology. Guide to Research Techniques in Neuroscience, 207–227. doi:10.1016/b978-0-12-374849-2.00009-4
- https://geneticeducation.co.in/gene-cloning-definitions-steps-procedure-applications-and-limitations/#Limitations_of_gene_cloning
- https://goldbio.com/articles/article/cloning-overview
- https://international.neb.com/tools-and-resources/feature-articles/foundations-of-molecular-cloning-past-present-and-future
- https://www.ncbi.nlm.nih.gov/books/NBK236044/
- https://www.ncbi.nlm.nih.gov/books/NBK26837/
- Lessard, J. C. (2013). Molecular Cloning. Laboratory Methods in Enzymology: DNA, 85–98. doi:10.1016/b978-0-12-418687-3.00007-0
- Sgaramella, V., & Bernardi, A. (2001). DNA Cloning. Encyclopedia of Genetics, 544–550. doi:10.1006/rwgn.2001.0350
- Watson J. D. Baker T. A. Bell S. P. Gann A. Levine M. & Losick R. M. (2004). Molecular biology of the gene (5th ed.). Pearson/Benjamin Cummings.
- Yeast Artificial Chromosome (YAC) (genosme.gov)
