Antibodies are often referred to as immunoglobulins (immune proteins).
Antibodies, or Y-shaped immunoglobulins, are proteins found in the blood that help to fight against foreign substances called antigens. Antigens, which are usually proteins or polysaccharides, stimulate the immune system to produce antibodies. The antibodies inactivate the antigen and help to remove it from the body. While antigens can be the source of infections from pathogenic bacteria and viruses, organic molecules detrimental to the body from internal or environmental sources also act as antigens. Genetic engineering and the use of various mutational mechanisms allow the construction of a vast array of antibodies (each with a unique genetic sequence).
Antibodies are all in a Y-shape with differences in the upper branch of the Y. These structural differences of amino acids in each of the antibodies enable the individual antibody to recognize an antigen. An antigen has on its surface a combining site that the antibody recognizes from the combining sites on the arms of its Y-shaped structure. In response to the antigen that has called it forth, the antibody wraps its two combining sites like a “lock” around the “key” of the antigen combining sites to destroy it.
An antibody’s mode of action varies with different types of antigens. With its two-armed Y-shaped structure, the antibody can attack two antigens at the same time with each arm. If the antigen is a toxin produced by pathogenic bacteria that cause an infection like diphtheria or tetanus, the binding process of the antibody will nullify the antigen’s toxin. When an antibody surrounds a virus, such as one that causes influenza, it prevents it from entering other body cells. Another mode of action by the antibodies is to call forth the assistance of a group of immune agents that operate in what is known as the plasma complement system. First, the antibodies will coat infectious bacteria and then white blood cells will complete the job by engulfing the bacteria, destroying them, and then removing them from the body.
There are five different antibody types, each one having a different Y-shaped configuration and function. They are the IgG, IgA, IgM, IgD, and IgE antibodies.
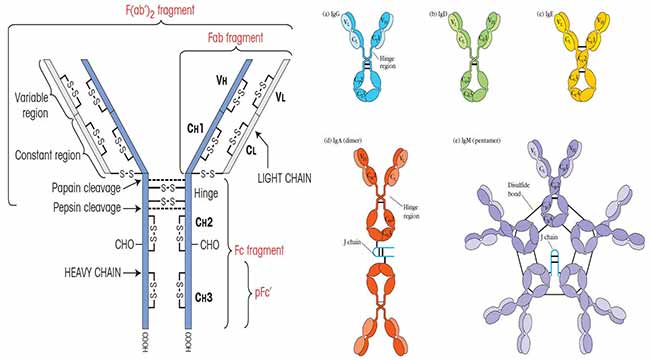
Image Source: Kuby Immunology 7th Edition.
Interesting Science Videos
Structure of Antibodies (Immunoglobulins)
All antibody molecules share the same basic structural characteristics but display remarkable variability in the regions that bind antigens.
Both heavy chains and light chains consist of aminoterminal variable (V) regions that participate in antigen recognition and carboxy-terminal constant (C) regions; the C regions of the heavy chains mediate effector functions.
Heavy chain and Light Chain
An antibody molecule has a symmetric core structure composed of two identical light chains and two identical heavy chains. Both the light chains and heavy chains contain a series of repeating homologous units, each about 110 amino acid residues in length, that fold independently in a globular motif that is called an Ig domain.
In the heavy chains, the V region is composed of one Ig domain, and the C region is composed of three or four Ig domains.
Each light chain is composed of one V region Ig domain and one C region Ig domain. The N-terminal half of light chains is thus referred to as the variable, or VL, the region of the light chain, and the less variable part of the sequence is termed the constant, or CL, region. The two major light chain constant region sequences are referred to as k (kappa) or λ (lambda) chains. As more light-chain sequences were generated, it became apparent that the lambda chain constant region sequences could be further subdivided into four subtypes—λ 1, λ 2, λ 3, and λ 4—based on amino acid substitutions at a few positions.
In humans, the light chains are fairly evenly divided between the two light-chain classes; 60% of human light chains are kappa whereas only 40% are lambda. In mice, the situation is quite different: Only 5% of mouse light chains are of the lambda light-chain type. All light chains have a molecular weight of approximately 22 kDa.
Variable regions are so named because their amino acid sequences vary among antibodies made by different B cell clones. The V region of one heavy chain (VH) and the adjoining V region of one light chain (VL) form an antigen-binding site. Because the core structural unit of each antibody molecule contains two heavy chains and two light chains, every antibody molecule has at least two antigen-binding sites.
The C region Ig domains are distant from the antigen-binding site and do not participate in antigen recognition. The heavy chain C regions interact with other effector molecules and cells of the immune system and therefore mediate most of the biologic functions of antibodies.
In addition, heavy chains exist in two forms that differ at their carboxy-terminal ends: one form of the heavy chain anchors membrane-bound antibodies in the plasma membranes of B lymphocytes, and the other form is found only in secreted antibodies. The C regions of light chains do not participate in effector functions and are not directly attached to cell membranes.
Disulfide bonds
Heavy and light chains are covalently linked by disulfide bonds formed between cysteine residues in the carboxy terminus of the light chain and the CH1 domain of the heavy chain. Non-covalent interactions between the VL and VH domains and between the CL and CH1 domains may also contribute to the association of heavy and light chains. The two heavy chains of each antibody molecule are covalently linked by disulfide bonds.
In IgG antibodies, these bonds are formed between cysteine residues in the CH2 domains, close to the region known as the hinge. In other isotypes, the disulfide bonds may be in different locations. Non-covalent interactions (e.g., between the third CH domains [CH3]) may also contribute to heavy chain pairing.
Hypervariable regions
Most of the sequence differences and variability among different antibodies are confined to three short stretches in the V region of the heavy chain and to three stretches in the V region of the light chain. These segments of the greatest diversity are known as hypervariable regions.
Complementaritydetermining regions (CDRs)
The hypervariable loops can be thought to resemble fingers protruding from each variable domain, with three fingers from the heavy chain
and three fingers from the light chain coming together to form the antigen-binding site. Because these sequences form a surface that is complementary to the three-dimensional shape of the bound antigen, the hypervariable regions are also called complementaritydetermining regions (CDRs).
Hinge region
Antibody molecules are flexible, permitting them to bind to different arrays of antigens. Every antibody contains at least two antigen-binding sites, each formed by a pair of VH and VL domains. Many Ig molecules can orient these binding sites so that two antigen molecules on a
planar (e.g., cell) surface may be engaged at once. This flexibility is conferred, in large part, by a hinge region located between CH1 and CH2 in certain isotypes. The hinge region varies in length from 10 to more than 60 amino acid residues in different isotypes.
Fab region and Fc region
The hinge region is particularly susceptible to proteolytic cleavage by the enzyme papain. Papain cleavage resolves the antibody molecule into two identical fragments that retain the antigen-binding specificity of the original antibody (Fab regions), and the remaining region of the molecule, which consists of the non antigen-binding portion. This latter region, which is identical for all antibodies of a given class, crystallizes easily and was thus called the Fc region (fragment crystallizable). Each Fab region and Fc region of antibodies mediates its own particular functions during an antibody response to an antigen. The Fab regions bind to the antigen, and the Fc region of the antigen-coupled antibody binds to Fc receptors on phagocytic or cytolytic cells, or to immune effector molecules. In this way, antibodies serve as physiological
bridges between an antigen present on a pathogen, and the cells or molecules that will ultimately destroy it. A family of Fc receptors exists; each Fc receptor is expressed on a different array of cells and binds to a different class of antibodies.
Immunoglobulins Classes (Types)
Using antibodies directed toward the constant region of immunoglobulins and amino acid sequencing of immunoglobulins derived from plasmacytoma tumor cells, investigators discovered that the sequences of the heavy-chain constant regions fall into five basic patterns. These five basic sequences have been named with Greek letters: μ (mu), 𝛿 (delta), γ (gamma), ε (epsilon), and α (alpha). Each different heavy-chain constant region is referred to as an isotype, and the isotype of the heavy chains of a given antibody molecule determines its class.
Thus, antibodies with a heavy chain of the μ isotype are of the IgM class; those with a 𝛿 heavy chain are IgD; those with γ, IgG; those with ε, IgE; and those with α, IgA. Th e length of the constant region of the heavy chains is either 330 amino acid residues (for γ, 𝛿, and α chains) or 440 amino acids (for μ and ε chains). Correspondingly, the molecular weights of the heavy chains vary according to their class. IgA, IgD, and IgG heavy chains weigh approximately 55 kDa, whereas IgM and IgE antibodies are approximately 20% heavier.
Physical Properties of Ig Classes
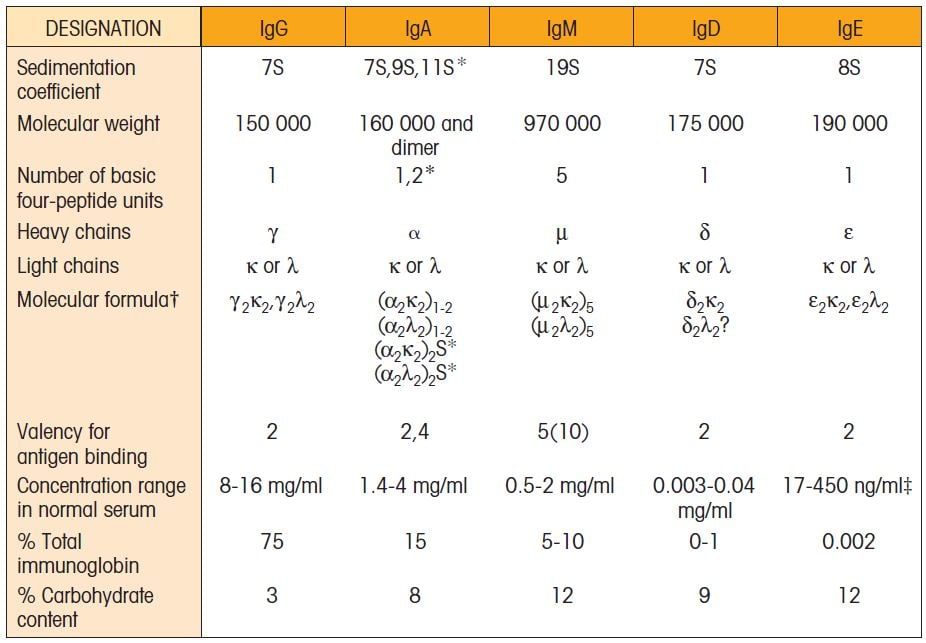
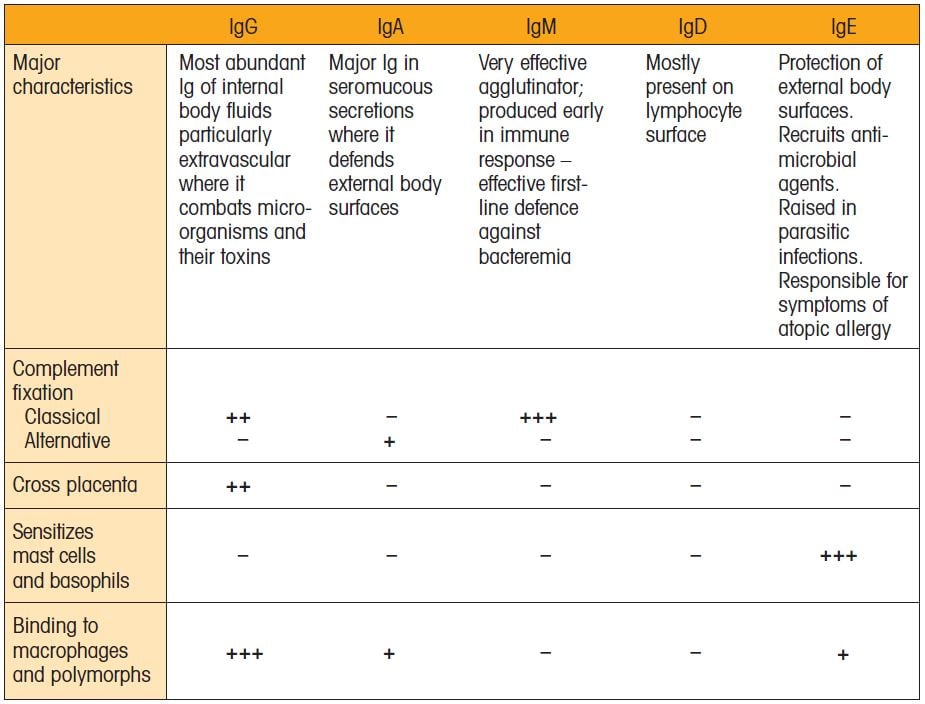
Biological properties of major Ig
Structures of the five major classes of antibodies
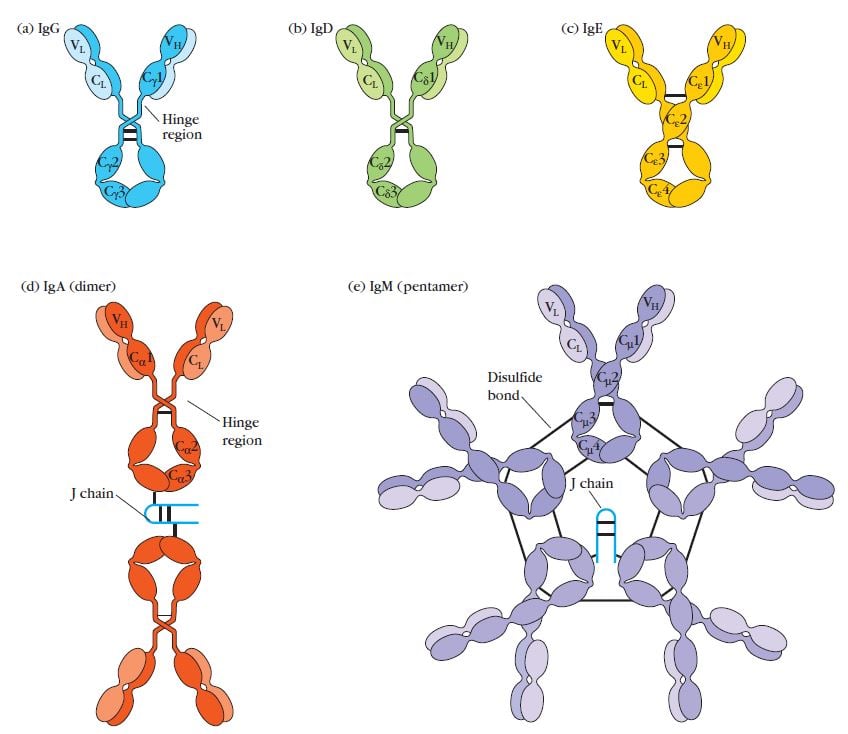
Figure: General structures of the five major classes of antibodies. Image Source: Kuby Immunology 7th Edition.

Hi, I would like to use your picture for my PhD thesis, is it possible to have the permission?
Thank you very much, I am looking forward to hearing from you
Well structured. An introduction yet complete. Good job. Keep it going. Congratulations!