Genomics is a field of science that focuses on the study of an organism’s entire set of genetic material, called its genome.
This includes studying the structure, function, and interactions between all of the genes and other components that make up the genome.
The genome of an organism is made up of DNA, which contains the instructions for how that organism develops, functions, and interacts with the world around it. By studying an organism’s genome, researchers can learn more about its traits, behaviors, and susceptibility to diseases.
Interesting Science Videos
Categories of Genomics
Two broad categories of genomic studies are – structural genomics and functional genomics.
- Structural genomics is the field of genomics that deals with structures of genome sequences. Understanding the genome structure involves constructing genome maps, sequencing genes, annotating gene features, and comparing genome structures.
- Functional genomics deals with the study of gene expression and the function of genes in a genome. It involves studying gene functions at the whole genome level using high-throughput methods.
In addition to structural and functional genomics, there are other subfields of genomics, including:
- Epigenomics involves the study of epigenetic modifications or epigenome, which refers to the collection of chemical compounds that attach to DNA and influence its activity. The epigenome plays an important role in determining the differences between various cell types in the body. Epigenomic modifications include DNA methylation and histone modification.
- Metagenomics involves the study of genetic material from entire biological communities rather than just individual organisms. It is generally applied to microorganisms. Metagenomics can be used to study the diversity and function of microbial communities in diverse environments, such as the human gut, soil, or ocean.
- Pharmacogenomics is the subfield of genomics that uses an individual’s genetic information to study and customize the choice and dosage of drugs in medical treatment. It can be used to predict drug response or toxicity and to develop a more personalized approach to prescribing drugs.
- Comparative genomics involves the comparison of genomes from different species that can provide insights into evolutionary relationships, functional elements, and genetic variations among species. It uses various tools that help to identify and understand the similarities and differences in the genomes of various species.
Methods in Structural Genomics
Genome mapping
- Genome mapping is a crucial process in structural genomics which involves the identification of the locations of genes, mutations, or traits on a chromosome.
- There are different types of genome maps, including genetic linkage maps, physical maps, and cytologic maps.
- Genetic linkage maps, also referred to as genetic maps, are low-resolution maps that use inheritance patterns to determine the relative positions of genetic markers on a chromosome.
- Physical maps are high-resolution maps that identify the locations of recognizable landmarks on a genomic DNA regardless of inheritance patterns.
- Cytologic maps, also known as chromosome maps, describe the banding patterns visible on stained chromosomes, which can be directly observed under a microscope. These visually distinct light and dark bands serve as markers on the chromosomes.
Genome sequencing
- The most detailed genome map is obtained through genome sequencing, which determines the complete DNA sequence of the genome.
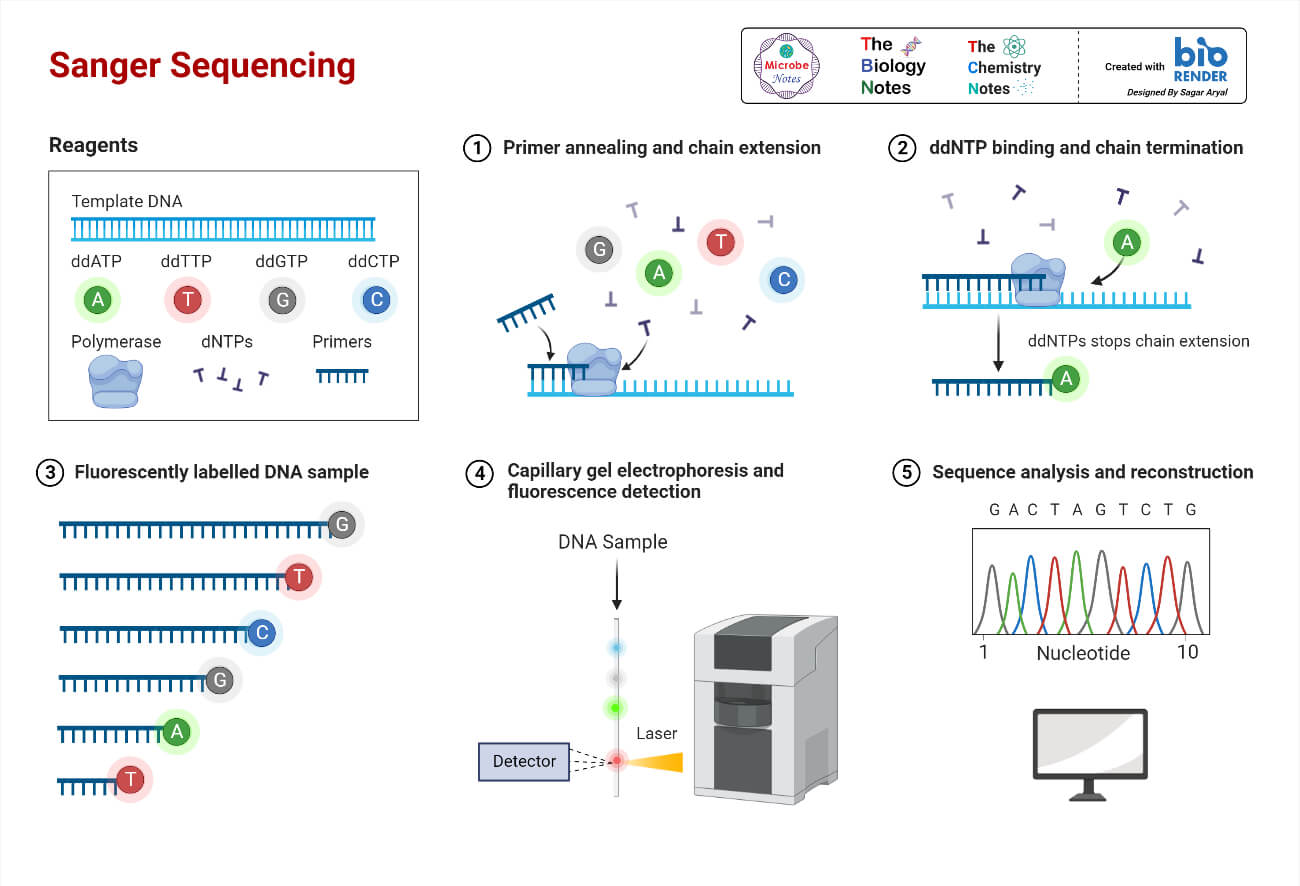
- A widely used DNA sequencing method is Sanger sequencing. It uses fluorescently labeled dideoxy nucleotides to terminate DNA chains of varying lengths. The resulting fragments are separated by electrophoresis, and the sequence is determined by reading the banding pattern in the gel. Base calling is used to assign bases for each peak in a chromatogram.
- Whole genome sequencing can be done using two main methods: shotgun sequencing and hierarchical sequencing.
- In shotgun sequencing, DNA clones are randomly sequenced from both ends. This generates a large number of DNA fragments, which are then assembled by a computer program to create a complete genome sequence.
- The hierarchical method involves mapping chromosomes using physical mapping techniques. Longer fragments of genomic DNA are cloned into a bacterial vector with high capacity. The physical map helps determine the locations and order of the cloned fragments on the chromosome.
Genome sequence assembly
- Genome assembly is the process of reconstructing a complete genome sequence from a large number of short DNA fragments called reads.
- DNA sequencing reactions produce short sequence reads derived from DNA clones. To reconstruct the complete genome, these short fragments need to be joined together while removing any overlapping regions.
- The first step in the genome assembly process is base calling which derives base calls and assigns quality scores to these base calls. Base calling is used to determine the sequence of nucleotides at every position in the DNA fragments. Quality scores are assigned to each base call, which represents the confidence or accuracy of the base call.
- The next step is to assemble the individual sequence reads into contiguous sequences, known as contigs. This involves identifying overlaps between the different fragments, determining their relative order, and then deriving a consensus sequence for each contig.
- There are several commonly used programs for processing raw sequencing data and assembling contigs, including Phred, Phrap, TIGR Assembler, and ARACHNE.
Genome annotation
- After the assembly of the genome sequence, it needs to be analyzed to identify and annotate useful biological features. The process of analyzing and interpreting genome sequences using computational methods is called genome annotation.
- Genome annotation process involves two main steps: gene prediction and functional assignment.
- In gene prediction, computational programs are used to predict the locations and structures of genes within the genome.
- The next step is to assign functions to the predicted genes. This is done through homology searching, primarily using BLAST.
- The resulting annotations are deposited into public databases like GenBank, making them accessible to the scientific community for further analysis and research.
Methods in Functional Genomics
Genetic interaction mapping
- Genetic interaction mapping is a technique used to study the functional relationships between genes. It refers to the study of how multiple genes interact with each other to influence specific phenotypic traits.
- This approach involves perturbing genes in pairwise combinations, such as through knockout, knockdown, or overexpression, to understand how one gene influences the phenotype of another.
- Epistasis is one such genetic interaction. It is a form of interaction between non-allelic genes that occurs when the effect of one gene is masked by another gene.
- There are numerous computational tools available for detecting gene-gene interactions. Some examples include BEAM (Bayesian Epistasis Association Mapping), TEAM (Tree-Based Epistasis Association Mapping), BOOST (Boolean Operation-based Screening and Testing), and TS-GSIS (Two Stage-Grouped Sure Independence Screening).
- Genetic interaction mapping is useful in discovering new gene functions and organizing gene products into functional complexes and pathways in a hierarchical manner.
Microarray technology
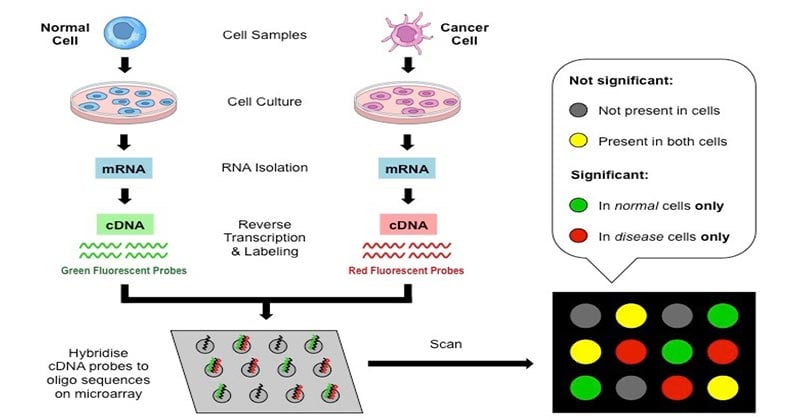
- Microarray-based approaches are widely used in genomics research for global gene expression profiling.
- A microarray is a chip that contains a high-density array of immobilized DNA oligomers or complementary DNAs (cDNAs). Each oligomer serves as a probe that can bind to a specific complementary cDNA molecule.
- In a microarray experiment, the labeled cDNA molecules are fluorescently or radioactively labeled and allowed to hybridize with the oligo probes on the chip. The amount of fluorescence or radiolabels at each spot on the microarray indicates the level of mRNA abundance in the cell.
- This technology enables the rapid and systematic analysis of gene expression patterns and facilitates the discovery of novel gene functions and regulatory mechanisms.
- Microarrays have various applications in functional genomics. They are used for comparing gene expression patterns between normal and diseased tissues, identifying single nucleotide polymorphisms (SNPs) in genes, and detecting alterations in chromosomal copy numbers.
SAGE
- SAGE (Serial Analysis of Gene Expression) is a high-throughput, sequence-based method used for analyzing gene expression profiles. It provides information about mRNA expression levels in a cell.
- Unlike microarray technology, which relies on hybridization, SAGE directly sequences RNA molecules to provide a global analysis of gene expression.
- In SAGE, short oligonucleotide sequence tags called SAGE tags are isolated from individual mRNAs and used as unique markers for gene transcripts.
- These tags are concatenated, cloned, and sequenced. The analysis of gene expression is performed computationally in a serial manner. The frequency of each gene tag indicates the level of gene expression.
- SAGE also has some limitations. The sequencing step is costly and time-consuming, and determining the number of tags to be sequenced for good coverage of the entire transcriptome can be challenging.
Applications of Genomics
- Genomics helps in the diagnosis and classification of diseases by identifying specific genetic changes. Genomics studies the genetic basis of diseases, identifying variations linked to specific conditions, predicting disease risk, and developing targeted treatments.
- Another application of genomics is pharmacogenomics which studies the impact of genes on an individual’s response to drugs. It analyzes genetic variations to determine the most effective and safe medications for personalized medicine.
- Comparative genomics compares the genomes of different species in order to identify similarities, differences, and evolutionary relationships. This helps in understanding the genetic basis of traits, evolutionary processes, and the diversity of life on Earth.
- Genomics also plays a crucial role in agriculture by enhancing crop and livestock breeding, improving disease resistance, and increasing agricultural productivity. It helps in the identification of genes related to desired traits and in developing superior crop varieties and livestock breeds.
- Genomic analysis also has applications in forensic and criminal investigations. DNA samples found at crime scenes can provide valuable information, help identify suspects and determine relationships between individuals.
References
- Bayón, G. F., Fernández, A. F., & Fraga, M. F. (2016). Bioinformatics Tools in Epigenomics Studies. Epigenomics in Health and Disease, 73–107. doi:10.1016/b978-0-12-800140-0.00004-2
- Chaitanya, K.V. (2019). Applications of Genomics. In: Genome and Genomics. Springer, Singapore. https://doi.org/10.1007/978-981-15-0702-1_8
- Clark, D. P., & Pazdernik, N. J. (2013). Genomics & Systems Biology. Molecular Biology, 248–272. doi:10.1016/b978-0-12-378594-7.00009-3
- Genetic Interaction Mapping | Krogan Lab (ucsf.edu)
- https://www.ebi.ac.uk/training/online/courses/functional-genomics-i-introduction-and-design/what-is-genomics/
- https://www.genome.gov/about-genomics/fact-sheets/A-Brief-Guide-to-Genomics
- https://www.genome.gov/about-genomics/fact-sheets/Comparative-Genomics-Fact-Sheet
- https://www.genome.gov/about-genomics/fact-sheets/Epigenomics-Fact-Sheet
- https://www.genome.gov/genetics-glossary/Pharmacogenomics
- https://www.ncbi.nlm.nih.gov/books/NBK569502/
- Kaur, S. (2013). Genomics. Brenner’s Encyclopedia of Genetics, 310–312. doi:10.1016/b978-0-12-374984-0.00642-2
- Kaushik, S., Kaushik, S., & Sharma, D. (2018). Functional Genomics. Reference Module in Life Sciences. doi:10.1016/b978-0-12-809633-8.20222-7
- Sivashankari, S., & Shanmughavel, P. (2007). Comparative genomics – A perspective. Bioinformation, 1(9), 376-378. https://doi.org/10.6026/97320630001376
- Xiong, J. (2006). Essential Bioinformatics. Cambridge: Cambridge University Press. doi:10.1017/CBO9780511806087
