DNA is the basic unit of inheritance that maintains the integrity and function of living organisms. However, it is constantly exposed to damaging agents which can cause DNA damage. Additionally, errors can occur during DNA replication and repair processes, leading to harmful mutations.
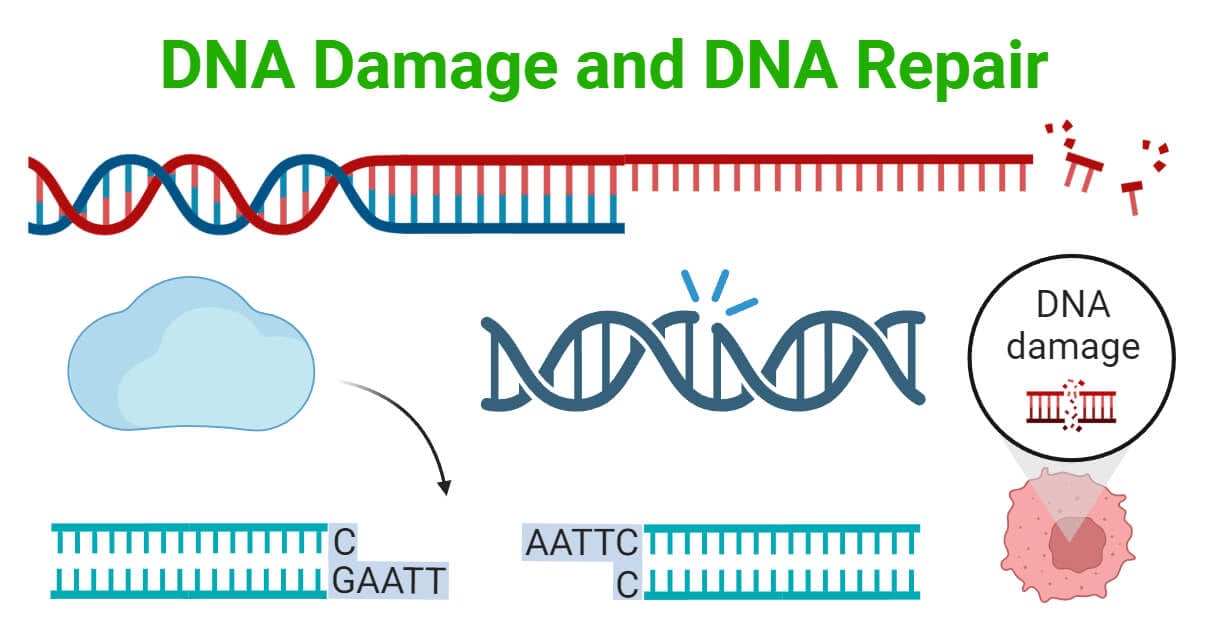
To overcome the harmful effects of DNA damage, cells have various systems such as DNA repair mechanisms, damage tolerance pathways, cell cycle checkpoints, and cell death pathways. These systems work together to repair or tolerate DNA damage, ensuring the overall survival and functionality of cells.
Understanding the mechanisms of DNA damage and the corresponding repair pathways is essential to understand the impact of DNA damage on cellular functions and the development of diseases.
Interesting Science Videos
What is DNA Damage?
DNA damage refers to changes or disruptions that occur in the DNA molecule caused by environmental factors or normal processes that happen inside our cells.
Types of DNA Damage and Mechanisms
There are various types of DNA damage that can happen due to normal cellular processes or exposure to damaging agents in the environment.
Several types of DNA damage are described as follows:
1. DNA Strand Breaks
DNA strand breaks occur when one or both strands of DNA are interrupted. There are two types: single-strand breaks (SSBs) where one strand is cut, and double-strand breaks (DSBs) where both strands are cut. These breaks can be caused by ionizing radiation like X-rays and gamma rays, as well as certain chemicals.
2. Oxidative Damage
Oxidative damage can occur due to the action of reactive oxygen species (ROS) which leads to the formation of lesions. The highly reactive ROS, such as hydroxyl radicals (•OH), can cause oxidative damage to DNA bases.
3. Alkylation of Bases
Alkylating agents, both endogenous and exogenous, can modify DNA bases by introducing alkyl groups. These modifications can be cytotoxic, mutagenic, or have neutral effects on the cell.
4. Base Loss
Base loss occurs when the nitrogenous bases in DNA are removed, leaving behind apurinic/apyrimidinic (AP) sites or abasic sites. AP sites are chemically unstable and can lead to DNA strand breaks or mutagenic events if left unrepaired.
5. Bulky Adduct Formation
Bulky adducts are formed when certain chemicals, such as polycyclic aromatic hydrocarbons (PAHs), covalently bind to DNA bases. These adducts create bulky modifications that stick out from the DNA and disrupt its structure. They can interfere with DNA replication, transcription, and repair processes, potentially leading to mutations.
6. DNA Crosslinking
DNA crosslinking occurs when two nucleotides in DNA become covalently linked together. Crosslinks can form within the same DNA strand (intrastrand crosslinks) or between opposite DNA strands (interstrand crosslinks). DNA crosslinks prevent the separation of DNA strands during replication or transcription, leading to the disruption of important cellular processes.
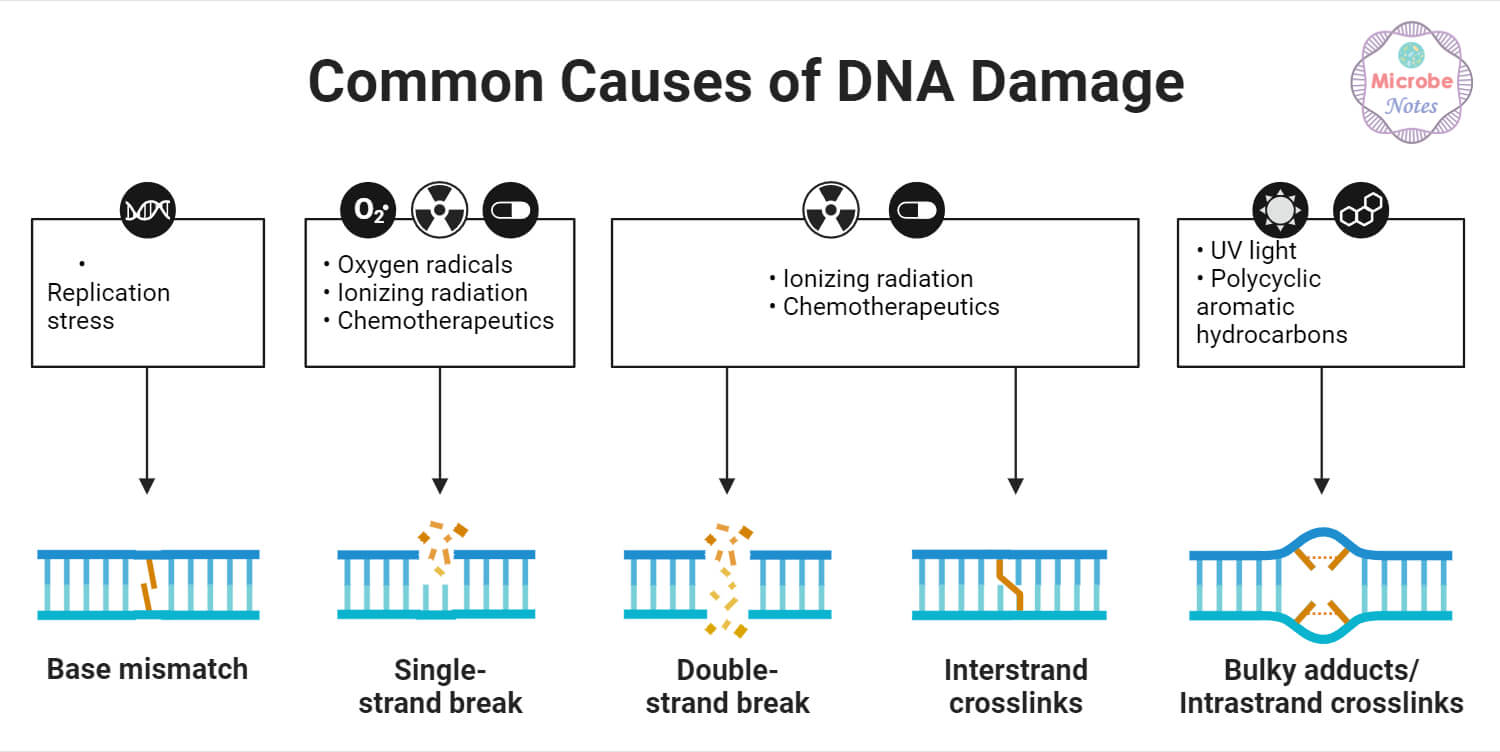
Sources/Agents of DNA Damage
DNA damage can also be classified into two types based on its origin or sources: endogenous and exogenous. The major sources of endogenous and exogenous DNA damage are briefly explained below:
1. Endogenous DNA Damage
Endogenous DNA damage originates from internal reactions involving chemically active DNA within cells.
- Replication errors are one source of endogenous DNA damage that occurs during DNA replication when incorrect nucleotides are inserted opposite the template bases. During replication, some DNA polymerases with lower fidelity can be involved, leading to potential errors.
- Topoisomerase enzymes are another source of endogenous DNA damage. Topoisomerases remove the supercoiling of DNA during replication and transcription. However, misalignment of the DNA ends can stabilize the topoisomerase-DNA cleavage complex and result in the formation of DNA lesions.
- Reactive oxygen species (ROS) are produced during cellular processes and can cause oxidative damage to DNA. While ROS plays an important role in normal cellular functions, excessive levels can lead to various DNA lesions and modifications. Excessive ROS has been associated with the development of several human diseases like cancer, Alzheimer’s disease, and diabetes.
- Alkylating agents are reactive compounds that can add methyl or ethyl groups to DNA bases, leading to chemical modifications. Spontaneous methylation events can generate different methylated bases. Some methylated bases are mutagenic and can lead to specific types of mutations.
2. Exogenous DNA Damage
Exogenous DNA damage is caused by external factors, such as environmental agents, physical forces, or chemicals.
- Ionizing radiation (IR) directly damages DNA or indirectly affects it through the generation of highly reactive hydroxyl radicals (•OH) from water molecules. IR can cause different types of damage to the DNA such as base lesions, and single-strand and double-strand breaks.
- Ultraviolet (UV) radiation is another agent of DNA damage. It is the leading cause of skin cancers. UV light can form pyrimidine dimers where two pyrimidines on the same DNA strand are joined together. This alteration in DNA structure can block transcription and replication processes.
- Exogenous alkylating agents, found in sources like tobacco smoke and industrial activities, react with DNA and can cause mutagenic and carcinogenic changes. They primarily target the nitrogenous bases in DNA. Examples of alkylating agents include sulfur and nitrogen mustards.
- Aromatic amines, found in cigarette smoke, fuel, coal, dyes, and pesticides, are also exogenous sources of DNA damage. These agents can create long-lasting lesions in the DNA structure that lead to the substitution of DNA bases and frameshift mutations.
- Polycyclic aromatic hydrocarbons (PAHs) are known carcinogens found in sources like tobacco smoke, automobile exhaust, and other environmental pollutants. PAHs require activation by the liver’s P-450 system to produce reactive substances that can potentially damage DNA.
What is DNA Repair?
DNA damage is a common event that can interfere with important cellular processes and lead to genomic defects and an increased risk of cancer. To ensure the integrity of their genomes, cells have evolved to develop mechanisms for DNA repair. These mechanisms help cells to cope with DNA damage.
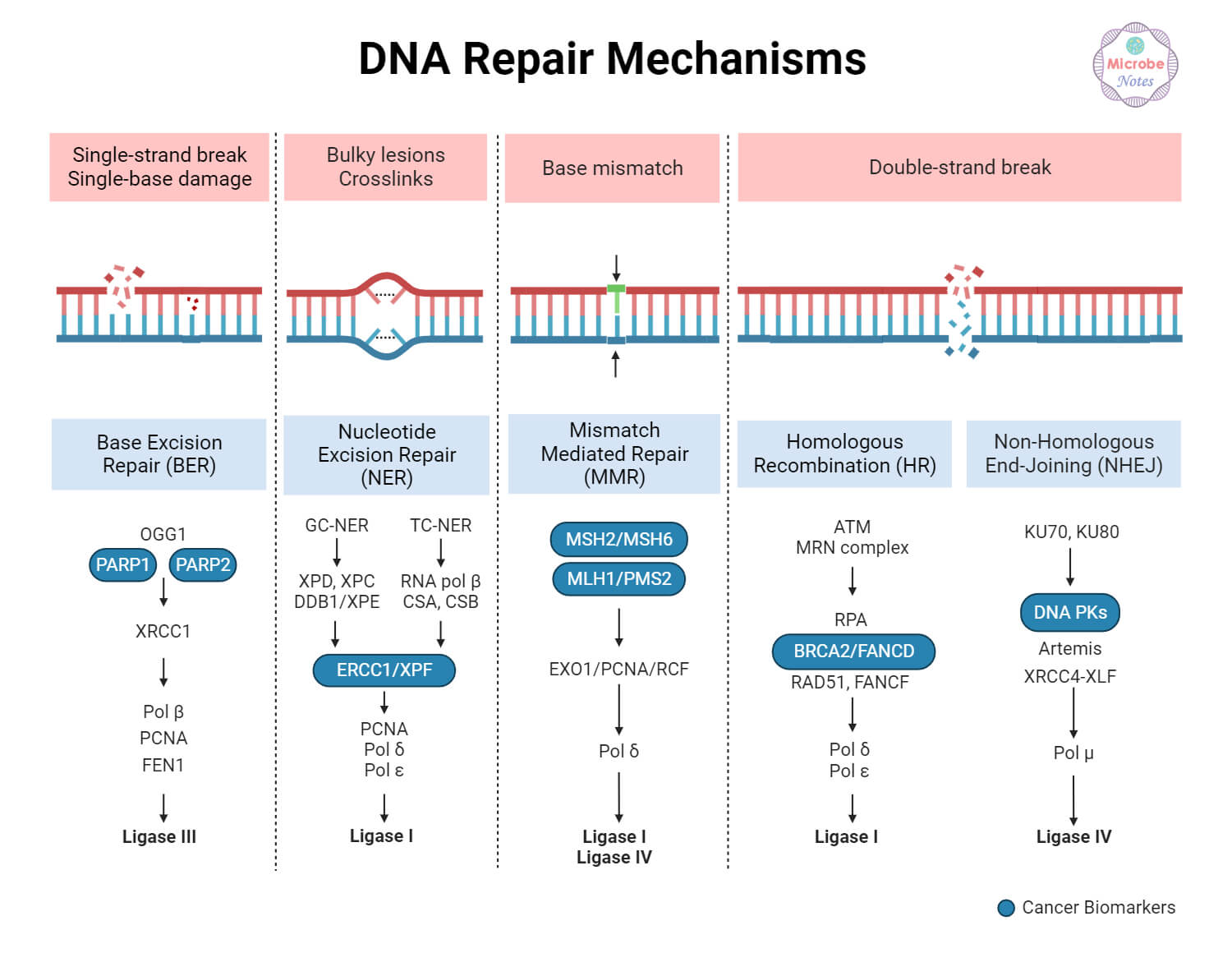
DNA Repair Types and Mechanisms
Various pathways exist for DNA repair. These include direct reversal, excision repair, mismatch repair, and repair of DNA breaks.
1. Direct reversal repair
- Direct reversal repair is a DNA repair mechanism that directly fixes specific types of DNA damage without the need for excision or replacement.
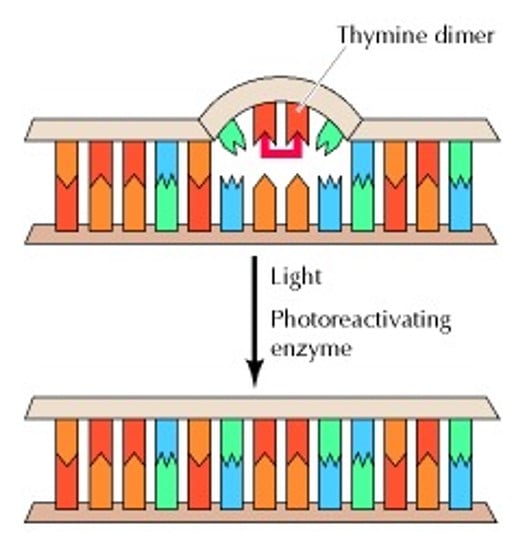
- Two examples of DNA damage that can be reversed are UV-induced lesions and alkylated bases.
- UV-induced lesions, caused by UV light, can be reversed through a process called photoreactivation, which uses visible light energy to break the damaged DNA structure, restoring the original pyrimidine bases.
- Alkylated bases can be reversed by enzymes such as O6-alkylguanine-DNA alkyltransferase (AGT) and AlkB-related dioxygenases, which removes or modifies the alkyl group, respectively.
2. Base excision repair
- Base excision repair (BER) is a DNA repair mechanism that removes and replaces damaged bases.
- It involves the action of various DNA glycosylases such as 8-oxoguanine DNA glycosylase (OGG1). These enzymes recognize and remove damaged bases.
- BER includes both short patch repair, where an abasic site is processed and filled by specific enzymes, and long patch repair, where gaps are tailored and DNA synthesis occurs followed by ligation.
- One example of BER is the repair of uracil-containing DNA. In this process, a DNA glycosylase recognizes and removes the uracil base, creating a gap in the DNA called AP site. The gap is then cleaved by an enzyme called AP endonuclease. After that, the remaining sugar is removed, and the gap is filled using DNA polymerase and sealed with ligase.
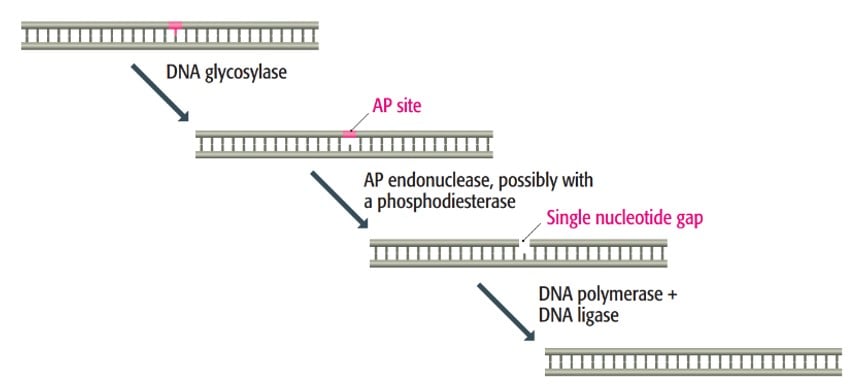
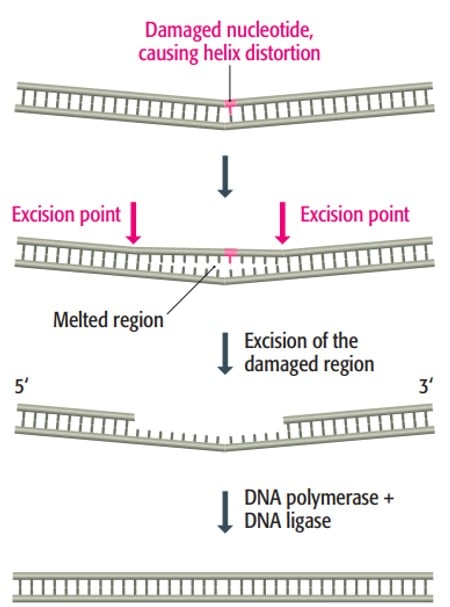
3. Nucleotide excision repair
- Nucleotide excision repair (NER) deals with bulky adducts and cross-linking lesions caused by UV radiation or chemical exposure.
- NER removes a fragment of nucleotides containing the damaged lesion and synthesizes a new DNA strand using the undamaged strand as a template.
NER consists of two pathways:
- Global Genome NER (GG-NER) repairs bulky damages throughout the entire genome, including regions that are not actively transcribed.
- Transcription-Coupled NER (TC-NER repairs damage that occurs on the transcribed DNA strand.
- Mutations in NER pathway genes can lead to disorders such as xeroderma pigmentosum (XP) and certain other neurodegenerative conditions.
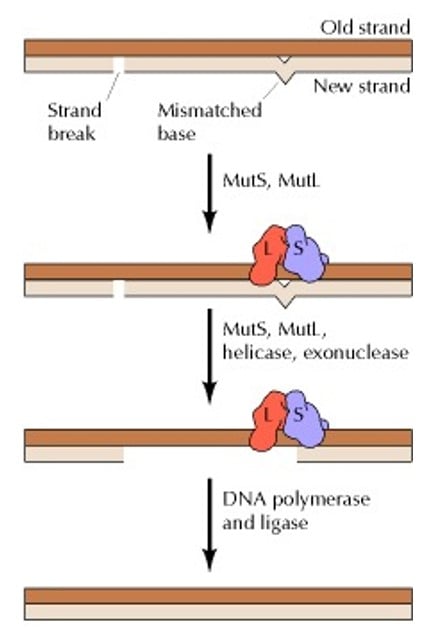
4. Mismatch repair
- Mismatch repair (MMR) pathway repairs base mismatches and insertion-deletion loops that occur during replication. Most of these errors are fixed by the proofreading activity of DNA polymerase during replication, but some may be missed and need to be corrected later.
- The MMR pathway involves three steps: recognition of mismatches, degradation of the error-containing strand, and synthesis of the correct DNA sequence.
- First, protein complexes such as MSH2-MSH6 in the MutS protein locate the mismatch errors and forms a complex with MutL which helps in further repair. MutS and MutL are important protein complexes in eukaryotes. In E. coli, another protein MutH also has an important role in mismatch repair.
- Next, exonuclease 1 (Exo1) degrades the error-containing strand while replication protein A (RPA) prevents further DNA degradation by binding to the exposed DNA.
- Then, DNA polymerase δ synthesizes the correct sequence. Finally, DNA ligase then seals any remaining nicks in the repaired DNA.
- Mutations in MMR genes can lead to Lynch syndrome, a hereditary condition associated with an increased risk of colon, ovarian, and other cancers.
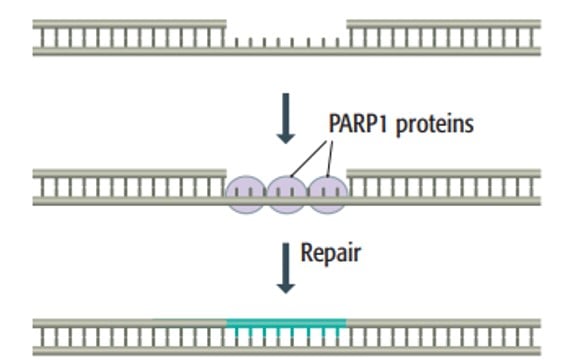
5. Single-strand break repair (SSBR)
- Single-stranded breaks (SSBs) in DNA can occur due to oxidative damage, abasic sites, or errors in the activity of the DNA topoisomerase enzyme.
- These breaks can disrupt DNA replication, halt transcription, and activate cellular processes that can lead to cell death.
- To protect the exposed single strand from breaking, PARP1 proteins coat the single strand and act as a shield.
- SSBR can be accomplished through various pathways already explained above, including base excision repair, nucleotide excision repair, and mismatch repair.
6. Double-strand break repair
Double-strand breaks (DSBs) in DNA can be repaired through two pathways: homologous recombination (HR) and non-homologous end joining (NHEJ).
Homologous recombination (HR)
- HR is a precise repair pathway that requires a matching DNA sequence as a template.
- It primarily uses the sister chromatid, a copy of the damaged DNA, for repair.
- HR is most active during the S, G2, and M phases of the cell cycle when sister chromatids are present.
- The HR process involves creating single-stranded DNA (ssDNA) by degrading one strand of the DNA break and coating it with proteins like RPA. Rad51 replaces RPA and pairs the ssDNA with a homologous DNA template for repair.
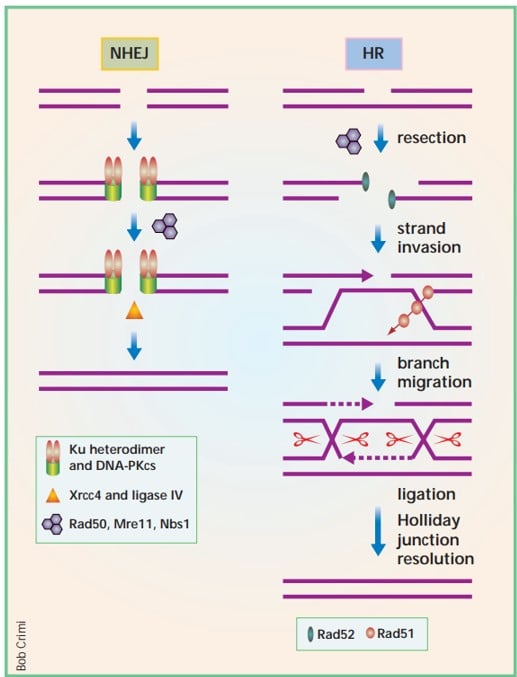
Non-homologous end joining (NHEJ)
- NHEJ is a simple and widely used mechanism that directly seals the broken ends of DNA without the need for a homologous DNA template.
- It can occur throughout the cell cycle.
- Proteins like Ku70/Ku80, DNA-PKcs, and LIG4/XRCC4 are involved in NHEJ. Ku70/Ku80 protects the DNA ends and prevents recombination, while DNA-PKcs and LIG4/XRCC4 help with end joining.
- The NHEJ pathway is faster but can be more error-prone compared to HR.
References
- 24.2: DNA Mutations, Damage, and Repair – Biology LibreTexts
- 5.13: DNA Repair – Biology LibreTexts
- Brown T. A. (2007). Genomes 3 (3rd ed.). Garland Science Pub.
- Chatterjee, N., & Walker, G. C. (2017). Mechanisms of DNA damage, repair and mutagenesis. Environmental and molecular mutagenesis, 58(5), 235. https://doi.org/10.1002/em.22087
- Cooper, G.M. (2000). DNA repair. In The cell: A molecular approach (2nd ed.). Sunderland, MA: Sinauer Associates. Retrieved from http://www.ncbi.nlm.nih.gov/books/NBK9900/.
- Dexheimer, T. S. (2012). DNA Repair Pathways and Mechanisms. DNA Repair of Cancer Stem Cells, 19–32. doi:10.1007/978-94-007-4590-2_2
- Hakem, R. (2008). DNA-damage repair; the good, the bad, and the ugly. The EMBO Journal, 27(4), 589-605. https://doi.org/10.1038/emboj.2008.15
- https://www.sigmaaldrich.com/NP/en/technical-documents/technical-article/research-and-disease-areas/cancer-research/dna-damage-and-repair
- Huang, R., & Zhou, P. (2021). DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduction and Targeted Therapy, 6(1), 1-35. https://doi.org/10.1038/s41392-021-00648-7
- Khanna, K. K., & Jackson, S. P. (2001). DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genetics, 27(3), 247–254. doi:10.1038/85798
- Vítor, A. C., Huertas, P., Legube, G., & de Almeida, S. F. (2020). Studying DNA Double-Strand Break Repair: An Ever-Growing Toolbox. Frontiers in Molecular Biosciences, 7, 512455. https://doi.org/10.3389/fmolb.2020.00024

Nice and informative article. Thanks Sanjuji.