Chromosome mapping means determining the relative positions of genes in a chromosome by creating maps that are used to organize and understand genetic information on chromosomes.
These maps show the positions of genes and the distances between them based on a specific scale. Chromosome mapping helps to understand the genome’s organization, structure, and function.
Thomas Hunt Morgan made significant contributions to the modern understanding of chromosome mapping. His experiments with fruit flies provided evidence for genetic linkage. Morgan’s discovery that the white eye gene in Drosophila was located on the X chromosome led to the identification and mapping of other X-linked genes. The resulting chromosome map revealed a linear arrangement of genes.
Alfred H. Sturtevant, an undergraduate in Morgan’s laboratory, invented the method for mapping chromosomes in 1911 by using the data from crosses with Drosophila and exploiting the phenomenon of genetic recombination. By studying the recombination frequency, he estimated the relative distances between genes on the chromosome. Sturtevant’s work established the foundation for studying gene organization on chromosomes.
Interesting Science Videos
Types of Chromosome Mapping
There are two main types of chromosome mapping techniques: Genetic Mapping and Physical Mapping.
1. Genetic Mapping
- Genetic mapping, also known as linkage mapping, refers to the method used to create genetic maps to estimate the positions of genes in chromosomes. These genetic maps are constructed by analyzing recombination patterns that occur during the crossing over of chromosomes.
- Genetic mapping starts by using linkage analysis to study the frequency of recombination between genes.
- Recombination frequency indicates genetic linkage and helps to determine whether the genes are linked together or not. A centimorgan (cM) is a unit that describes a recombination frequency of 1%.
- As the distance between two genes increases, the likelihood of recombination occurring also increases, resulting in a higher recombination frequency between them.
- The recombination frequency is determined by using the number of recombinant individuals compared to the total number of offspring. This frequency is expressed as a percentage.
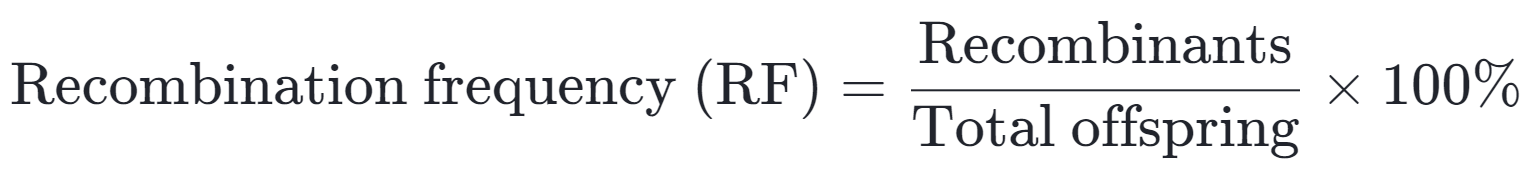
- It is important to understand the concept of genetic linkage and recombination for genetic mapping.
Genetic Linkage and Recombination
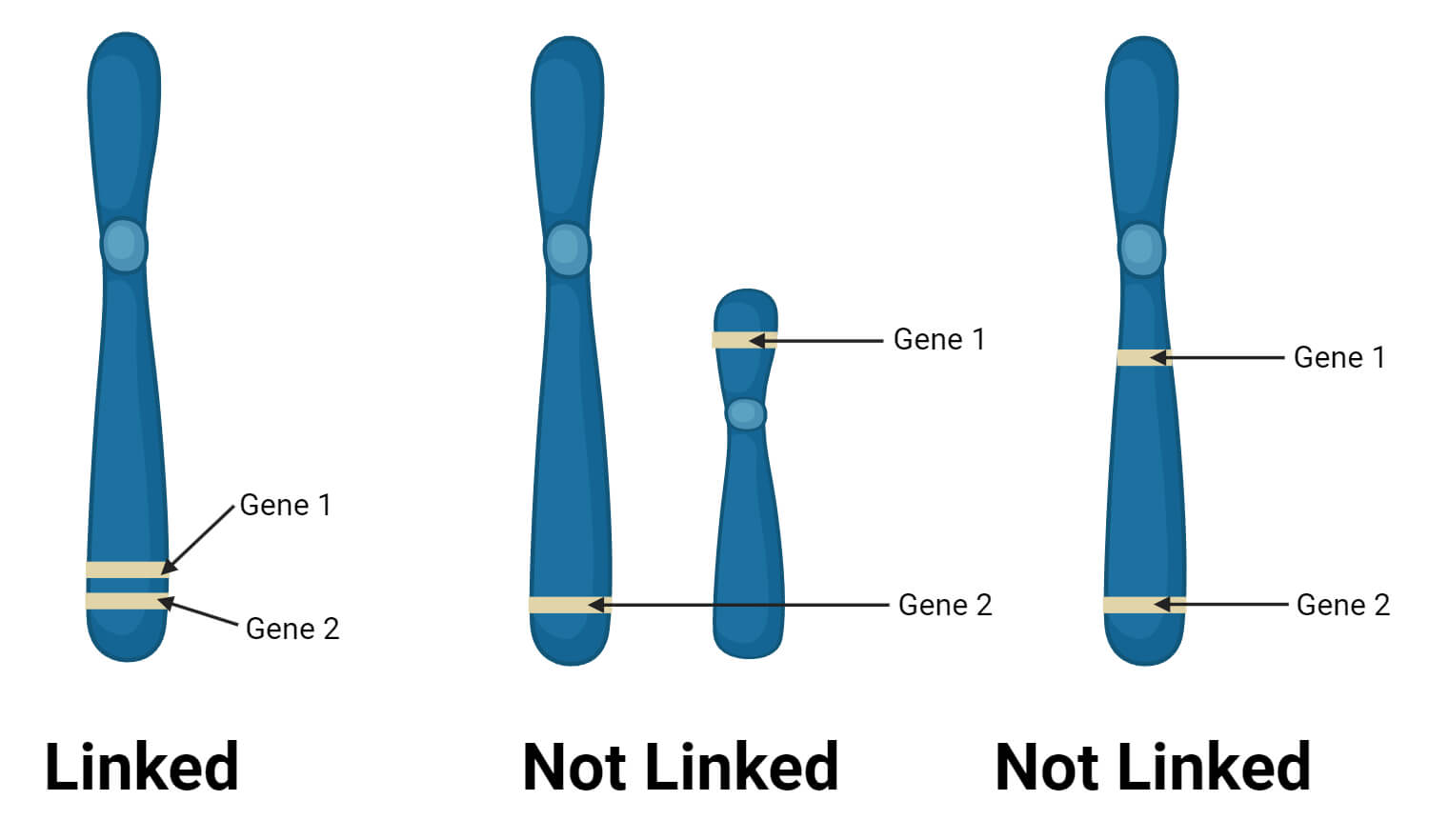
- Mendel’s law of independent assortment states that pairs of alleles segregate independently. However, upon the rediscovery of Mendel’s work, it was known that some pairs of genes were inherited together due to their location on the same chromosome. This phenomenon is known as genetic linkage.
- Linked genes are located close to each other on a chromosome, which increases the chance of them being inherited together. So, the concept of linkage is used to understand the relative positions of genes on a chromosome.
- However, not all genes on a chromosome are necessarily linked. Genes that are located further apart from each other have a higher chance of being separated during recombination.
- Recombination occurs during meiosis, where homologous chromosomes pair up to form bivalents, and crossing-over can occur between chromatids, leading to the exchange of genetic material.
- Genes that are closer together have a lower chance of crossover. Thus, the crossover percentage reflects the distance between the genes.
Molecular Markers for Genetic Mapping
- A molecular marker is a specific DNA sequence that shows variation and can be identified using molecular techniques. It is used in mapping to locate its position on the genome.
- To map molecular markers, their inheritance patterns are observed in subsequent generations using techniques like Southern hybridization and PCR.
- Data is collected from the analysis of molecular markers, including information about the presence or absence of specific markers.
- Using specialized software, the collected data is processed to construct a genetic map that shows the relative positions of markers on the genome.
Three commonly used molecular markers in genetic mapping are:
- Restriction Fragment Length Polymorphisms (RFLPs) are one of the earliest molecular markers used for genetic mapping. RFLPs are specific patterns of DNA fragments created by restriction enzymes that identify and cut DNA at specific sites. In genomic DNA, there can be variations in these restriction sites, leading to differences in the restriction patterns between closely related genomes. RFLP can be mapped on a genome by studying its inheritance pattern.
- Simple Sequence Length Polymorphisms (SSLPs) are DNA markers that consist of repetitive sequences with variations in the number of repeats. There are two types of SSLPs: minisatellites and microsatellites.
- Minisatellites are also known as the variable number of tandem repeats (VNTRs) and they have longer repeat units, up to 25 base pairs
- Microsatellites, also known as simple tandem repeats (STRs), have shorter repeat units, usually dinucleotide or tetranucleotide units.
- Single Nucleotide Polymorphisms (SNPs) are variations in a genome with different nucleotides at specific positions. SNPs can be detected using oligonucleotide hybridization, which allows selective pairing between an oligonucleotide and a DNA molecule. Techniques like DNA chip technology and solution hybridization have been developed for SNP screening, which offers faster and more efficient detection than gel-based methods.
Limitations of Genetic Mapping
- Genetic mapping does not provide information about the physical distance between genes as it provides only the outline.
- Recombination levels can differ across various parts of the genome. Some parts exhibit high rates of recombination, known as recombination hotspots, which can impact the accuracy of genetic mapping.
- In organisms where obtaining a large number of progenies is difficult, the resolution of a genetic map is limited because it depends on the number of observed crossovers.
- So, it is important to consider mapping information generated using other methods as well.
- Various physical mapping methods have been developed to address these limitations.
2. Physical Mapping
- Physical mapping involves determining the precise locations of DNA sequences on chromosomes. It uses base pair as the unit of measurement.
- Physical maps provide a direct representation of the physical structure of a chromosome and the positions of genes along its DNA sequence.
- Markers used in physical mapping include ESTs (Expressed Sequence Tags), STS (Sequence Tagged Site) markers, and genome-wide DNA sequences.
Methods of Physical Mapping
- Cytogenetic Mapping is a method used to locate genes on chromosomes by using their unique banding patterns. Each chromosome has its own distinctive pattern of light and dark bands when observed under a microscope. These bands are numbered to help in the precise identification of specific regions within a chromosome. Cytogenetic maps can be created by correlating the position of a gene with the corresponding band on the chromosome. These maps are also called idiograms.
- Restriction Mapping is a physical mapping method used to identify the positions of restriction sites on a DNA fragment. Restriction mapping involves comparing the sizes of DNA fragments created by two different restriction enzymes. Using RFLPs as markers for genetic mapping helps us to find the locations of certain variable restriction sites in a genome. However, most restriction sites are not variable, so RFLPs cannot be used to map many sites. Restriction mapping can be used as an alternative method in order to locate non-variable restriction sites.
- Fluorescence in situ hybridization (FISH) is a method used to visualize the position of specific DNA sequences on chromosomes. It involves labeling DNA probes and hybridizing them into intact chromosomes. In early versions of FISH, radioactive probes were used, but later fluorescent DNA labels were developed, which offer improved sensitivity and resolution.
- Sequence Tagged Site (STS) Mapping is a powerful mapping method for creating detailed physical maps of large genomes. It involves using short DNA sequences called STSs, typically 100 to 500 base pairs long, that occur uniquely in the genome. The map can be created by identifying the fragments that contain specific STSs using hybridization analysis or PCR. STSs can be obtained from various sources, including expressed sequence tags (ESTs), SSLPs, or random genomic sequences obtained through sequencing cloned genomic DNA. A commonly used marker for STS mapping is the EST, which is a unique sequence obtained by partially sequencing clones from a cDNA library.
- Radiation Hybrid (RH) Mapping is another physical mapping method that estimates the distance between genetic markers. RH mapping uses radiation to break DNA fragments. By controlling the radiation exposure, this method can create breaks between linked markers, resulting in a more detailed map. RH mapping is valuable for locating various genetic markers and genomic fragments, particularly in regions with limited availability of highly polymorphic markers.
Limitations of Physical Mapping
- DNA fragments can be incorrectly mapped due to fragment breakage, deletion during replication, or contamination with host genetic material.
- There may be missing or incomplete coverage of DNA fragments in the mapping process, resulting in gaps.
- Restriction mapping method cannot be applied to large genomes.
- Physical mapping using FISH is difficult to carry out and the process of accumulating data is slow. In a single experiment, only a limited number of map positions can be obtained.
- To address these limitations and improve overall mapping accuracy, it is important to integrate and cross-reference both physical and genetic mapping.
Importance of Chromosome Mapping
- Chromosome mapping is important for understanding the organization and arrangement of genes on chromosomes.
- Chromosome mapping is important for identifying the precise positions of genes on chromosomes, which can help to understand gene interactions.
- Chromosome mapping is also useful to understand the structure and function of genes.
- Chromosome mapping provides information about the location of genes which is important in the study of genetic disorders.
- Chromosome mapping is also useful for understanding evolution by comparing slight variations in gene arrangements among related species.
References
- 17.2: Mapping Genomes – Biology LibreTexts
- Griffiths, A. J. F., Miller, J. H., Suzuki, D. T., Lewontin, R. C., Gelbart, W. M. (2000). An introduction to genetic analysis (7th ed.). New York: W. H. Freeman.
- Hartl D. L. (2014). Essential genetics: a genomics perspective (6th ed.). Jones and Bartlett.
- http://www.cyto.purdue.edu/cdroms/cyto6/content/primer/mapping.htm
- http://www.informatics.jax.org/silver/chapters/7-1.shtml
- https://learn.genetics.utah.edu/content/pigeons/geneticlinkage
- https://www.genome.gov/about-genomics/fact-sheets/Genetic-Mapping-Fact-Sheet
- https://www.khanacademy.org/science/ap-biology/heredity/non-mendelian-genetics/a/linkage-mapping
- https://www.ncbi.nlm.nih.gov/books/NBK21116/
- https://www.ncbi.nlm.nih.gov/books/NBK218246/
- https://www.ncbi.nlm.nih.gov/books/NBK218246/
- O’Connor, C. (2008) Chromosome mapping: Idiograms. Nature Education 1(1):107
- Saraswathy, N., & Ramalingam, P. (2011). Genome mapping. Concepts and Techniques in Genomics and Proteomics, 77–93. doi:10.1533/9781908818058.77
- Speed, T. P., & Zhao, H. (2004). Chromosome Maps. Handbook of Statistical Genetics. doi:10.1002/0470022620.bbc01
- Williams, K. L. (2018). Gene Mapping. Reference Module in Life Sciences. doi:10.1016/b978-0-12-809633-8.20233-1

what about the linkage and recombination rules