Molecular docking is a computational approach used to predict the binding of molecules, referred to as ligands, to appropriate receptor proteins. It is a tool utilized for various computational purposes, including drug discovery. It has become an essential tool in in silico drug development. It is used to study ligand-receptor binding interactions using measures such as docking scores, g-scores, binding free energies, etc. Such a method allows researchers to study small molecules and ensure that they can yield beneficial results before taking them for wet lab experiments.

Molecular docking facilitates the analysis of interactions between two compounds, thereby enabling the creation of a stable complex that can be utilized in various experiments. The results from the docking study can then be used for energy profiling, strength, and stability of the complexes. The ligand utilized can be any small molecule; the target protein may be a protein, carbohydrate, or nucleic acid. The data collected from the docking studies can be stored as raw data in databases for future experiments and uses.
What is Molecular Docking?
Molecular docking is a computational method that has found its importance within the life science sector over the past three decades. The emergence of molecular docking was driven by the needs of structural molecular biology and structure-based drug discovery (referred to as rational drug discovery).
It is a computational modelling technique that facilitates the prediction of the binding orientation of a small molecule, commonly referred to as a ligand, to another biomolecule, referred to as the receptor protein. The molecular docking technique works by stabilising the structures and then studying their interaction with each other.
Most of the molecular docking tools available within the market contain built-in tools for the stabilisation of the structures and provide information about their binding free energy (∆Gbind), which is modeled in terms of dispersion & repulsion (∆Gvdw), hydrogen bond (∆Ghbond), desolvation (∆Gdesolv), electrostatic (∆Gelec), torsional free energy (∆Gtor), final total internal energy (∆Gtotal), and unbound system’s energy (∆Gunb).
Molecular docking has diverse uses and applications in drug discovery and development. This includes structure studies, lead (the drug candidate) optimization, screening for potential leads, predictions for mutagenesis studies, x-ray crystallography studies, chemical activity studies, etc. Molecular docking provides three-dimensional structural hypotheses of how a ligand would interact with its target.

Principle of Molecular Docking
To perform molecular docking, the first step is to search the structural data bank for a target of interest and a methodology to evaluate the ligand. For this evaluation, various molecular docking tools and methodologies are available. Such an evaluation allows for placing the ligands in a hierarchical order to ensure the best ligand that can interact with the target of interest. To establish the best interaction, an imaginative sampling of possible poses of ligands in the specified groove or pocket of the target is required to obtain the optimal binding geometry, which then decides the best interaction. Such an evaluation is done by scoring functions of docking software; however, X-ray crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy are the primary techniques for the investigation and establishment of three-dimensional structure data for biomolecular targets.
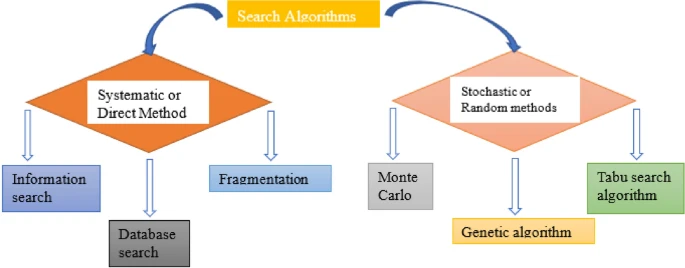
Scoring Functions
Scoring functions are mathematical models utilized to evaluate the interactions between the ligand and the receptor protein and establish a proper rank based on these interactions. Scoring functions are of three types: empirical, force field-based, or knowledge-based. In molecular docking, along with a scoring function, a search method is employed to explore state variables.
Scoring Functions Types
Systematic search method
Also referred to as the direct method. This method samples the search space at predefined intervals and is deterministic. This can be further divided into three subclasses:
- Conformational search: The flexibility of the ligand is achieved by rigidly docking an ensemble of regenerated ligand conformations with other programs (e.g., OMEGA). Then the ligand binding modes from other runs are ranked based on the binding energy scores. The ligand’s structural parameter is converted based on the torsional (dihedral), translational, and rotational degrees of freedom.
- Fragmentation: The basic idea behind this approach is that the ligand is first divided into a number of fragments. Multiple fragments can be docked to form bonds between them, or these fragments can be anchored separately. In the latter case, the first fragment is docked first, and subsequent fragments are built outward in steps from that initial bound position. It utilizes tools like Flex XTM, DOCK, LUDI, etc.
- Database Search: Also referred to as an exhaustive search. Here, all reasonable conformations of every ligand that are recorded in the databases are docked. This is quite uncomplicated; however, it produces a large pool of samples. This is because the flexible ligand docking is performed by systematically rotating all possible rotatable bonds of the ligand at a given interval. Thus, a filtration process is required to choose ligand conformations, which are then further subject to the more precise refinement and optimization measures. An example of a tool used is FLOG.
Stochastic search methods
Also referred to as random methods. Here, random changes are made to the state variables until a user-defined termination criterion is met, so the outcome of the search varies. This method is also divided into three subtypes:
- Monte Carlo: In such a method, ligands are placed in the receptor binding site, which is then cored, and a new configuration is generated. It employs instruments such as MCDOCK, ICM, etc.
- Genetic algorithm: The configuration and the location of the receptor molecule are described by the “gene,” and the score is “fitness”. The fit ligand receptor complex (poses) is then utilized to produce the next generation. It utilises programs like GOLD, AutoDock, and others.
- Tabu search: It works by setting constraints that enable the search for a new arrangement while avoiding re-examination of previously examined sections of the ligands’ conformational space. The tools it uses are PRO LEADS, Molegro Virtual Docker (MVD)TM, etc.
Search methods are also classified by the sample pool. It can either be local or global. The former is more focused on finding the nearest or local minimum energy to the current conformation, while the latter searches for the best or global minimum energy within the defined search space. Hybrid search methods can also be performed, which gives better results with lower energies.
Key Steps in Molecular Docking
Docking involves finding the most favorable binding mode(s) of a ligand to the target of interest; thus, the favorable ligand mode is obtained by the above-mentioned scoring function and search methods. Once that is done, the next steps are:
- Target Selection and Preparation
- Ligand Selection and Preparation
- Docking
- Evaluating Docking Results
1. Target Selection and Preparation
The target chosen should have appropriate conformation and be experimentally validated, preferably should have been validated by either X-ray crystallography or nuclear magnetic resonance. The targets can be taken from any online databases that contain experimentally validated molecules. Once the required target is chosen, the preparation is done. Most of the docking software available in the market has built-in tools required for target preparation. Here, the structure is selected and the binding sites to be interacted with. Once this is done, hydrogens must be added, with some programmed being more position-sensitive than others. In order to relax the structure and eliminate any steric interference, the protein is then subjected to energy minimization. The protonation states of ionizable residues in the protein are then determined, which is done to provide appropriate electrostatic interactions during docking. Water molecules and unnecessary ligands are removed from the protein structure to further simplify the system. Finally, the protein is supplied with the relevant force field parameters to accurately depict its behavior during docking simulations. The protein is then taken to the next step.
2. Ligand Selection and Preparation
Similar to the previous step, the ligand is selected from the database. The ligand chosen differs with lead optimization, lead discovery, and focused lead optimization. For each, certain filters should be fulfilled, and then chosen. Once chosen, the ligand is prepared. Here, the pKa values for each charged atom are predicted, and a program is implemented for each possible charge arrangement within a specified pH range. The chemical structure of the ligand can be reduced with the use of quantum mechanical force fields.
3. Docking
Before docking the ligand to the target protein, the active site of the target protein must be determined. This site is the binding pocket of the protein where the ligand can form interactions, which then lead to conformational changes. Once this is done, a search space is explored computationally, and candidates are ranked to determine the best binding mode. This is done by the aforementioned search method and a scoring function. In molecular docking, the scoring function and the search method go hand in hand.
4. Evaluating Docking Results
This is the final, yet the crucial step. Despite the ligand–protein docking tool used, the docking results are calculated, which provide information on the chemical complementarity between the ligand and the protein. The results should be evaluated to check whether all the necessary elements are fulfilled, such as the hydrogen bond donors and acceptors in the ligand, interaction of the charged groups in the ligand with oppositely charged side chains in the receptor, if the hydrophobic groups in the ligand are buried in hydrophobic pockets in the receptor, etc. Evaluation of the result can be done by calculating the binding affinity energy, which is alternatively calculated based on the predicted interaction energy. The ligands are then ranked based on their affinity scores. All this information can be useful for future reference and optimization.

Types of Molecular Docking
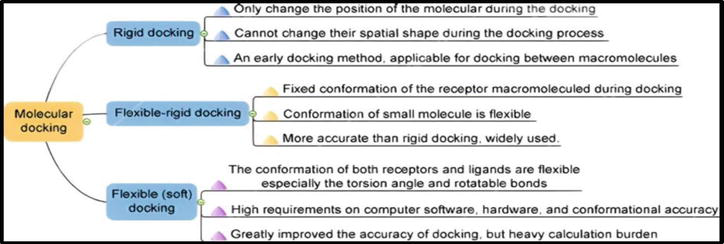
Molecular docking is of three kinds. These include:
- Flexible ligand docking: Most commonly used types in docking. Here, the target protein is incorporated as a rigid molecule.
- Rigid body docking: Here, both the target and ligand are kept as rigid molecules.
- Flexible docking: Both the interacting molecules, i.e., the ligand and target, are flexible.
Popular Molecular Docking Tools
There are multiple docking software available in the market. Some of the most commonly utilised ones are as stated below:
GOLD
Known as Genetic Optimisation for Ligand Docking, it was developed by the Cambridge Crystallographic Data Centre. It provides highly reliable results as it utilises genetic algorithms. It shows flexibility in handling diverse protein-ligand complexes, high accuracy, and allows the optimization of the scoring function. This software utilises numerous ligand subgroups. It shows a 71% success rate in determining the experimental binding mode for 100 protein complexes.
Website: https://www.ccdc.cam.ac.uk/solutions/software/gold/
AutoDock
This software is well-renowned for its power and flexibility. It provides impressive docking simulations and virtual screening. Its robustness, precision, and versatility make it a popular software for docking use.
Website: https://autodock.scripps.edu/
Flex-X
Compared to the aforementioned software, this is comparatively fast while providing accurate results. It allows users to construct ligand complexes incrementally and accounts for side-chain flexibility. It is suitable for high-throughput virtual screening.
SwissDock
It is a web tool dedicated to protein-small molecule docking. Perfect for beginners due to its user-friendly interface.
Website: https://www.swissdock.ch/
Some of the other popular software are Hammerhead, ICM, MCDock, GOLD, GemDock, Glide, and Yucca.
Models of Molecular Docking
The different models of molecular docking are:
- The lock and key theory
- The induced-fit theory
- The conformation ensemble model
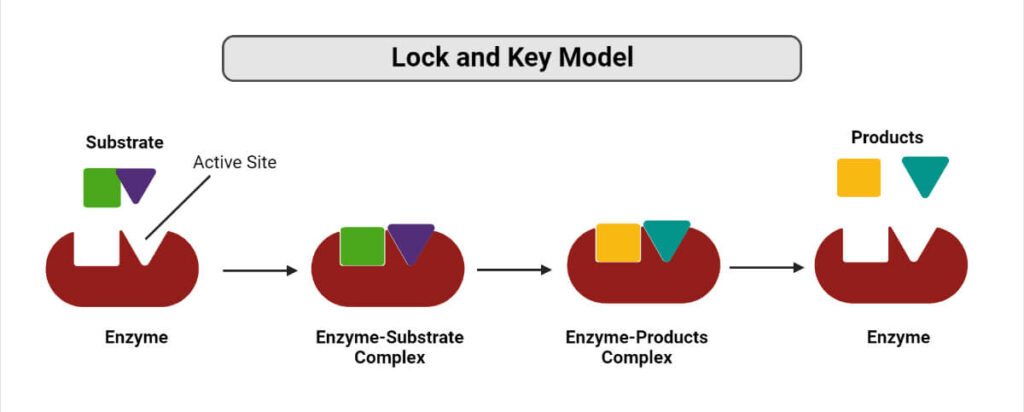
1. The lock and key theory
Emil Fischer, in 1890, described how biological processes operate with the concept of ‘The lock and key theory’. The principle behind this theory is that a substrate is inserted into the active site of a macromolecule in the same way as a key is inserted into a lock. The substrates exhibit distinct stereochemical properties that are required for their operation.
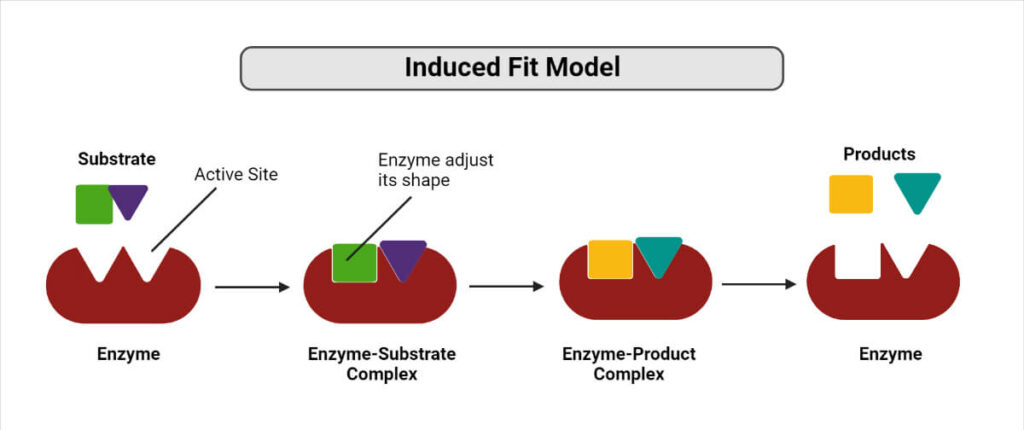
2. The induced-fit theory
Proposed by Daniel Koshland in 1958, this theory states that both the ligand and target adapt to one another by modest conformational changes until an ideal match is reached.
3. The conformation ensemble model
This model explains that proteins are composed of pre-existing ensembles of conformational states, and their flexibility enables them to transition between states. Proteins undergo significantly greater conformational changes.
File Formats in Molecular Docking
Depending on the docking programs used, file formats for receptors nd ligands become specific. File format offers a standardized way to represent the ligands and receptor proteins on a molecular level. This ensures that, despite the software utilities, there is a harmonization of different molecular docking software. Some of the file formats that are used are:
MOL2 (Tripos Mol2)
A Tripos Mol2 file (. mol2) is an ASCII file that contains all the required information to build a SYBYL molecule. This file format is made in a free file format, making it different from the fixed files. This makes it easily convertible to another file format. All the structural information of the molecule, such as the three-dimensional coordinates of each atom, atom types, types of bonds, partial charges, etc, is described by the MOL2 files.
SDF (structured data file)
A structured data file, developed by Biovia (previously Molecular Design Limited (MDL), is a sort of chemical data file format. This format provides two-dimensional and three-dimensional structural information about the ligands in plain text. SDF files, unlike MOL2 files, provide information about single and multiple ligands separated by the sign of four dollars ($$$$). It also encodes information about the connection and hybridization state. This file format is widely used for representing ligand structures, particularly in virtual screening investigations.
PDB (Protein Data Bank) file format
The Protein Data Bank (PDB) format is a standard for files that contain atomic coordinates. The structures from the Protein Data Bank use this file format, which can be read and written by a variety of tools. The whole PDB file specification contains a plethora of information, including authors, literature references, and the process of structure determination. PDB format is made up of lines of information in a text file, and these lines are referred to as a record. A PDB file often contains several different sorts of records.
PDBQT (Protein Data Bank, Partial Charge, and Atom Type)
This is an extension of the PDB file format. However, it’s a bit more descriptive, containing information about the ligands’ partial charges (q) and atom types (t). The partial charges are required for the calculation of electrostatic interactions between the molecules during docking carried out by AutoDock. PDBQT format also contains information on the rotatable bonds to depict the flexible compounds, allowing for the docking software to investigate the conformational space of the ligand.
XYZ (Cartesian coordinates)
Compared to other file formats, the XYZ file format is simple as it represents the total ligand atoms in the first row and a commentary in the second row. The three-dimensional coordinates of the ligands are then listed from the third row. These ligands are represented as Cartesian (X, Y, and Z) coordinates. These files can be generated in some docking software quite easily.
Use of Artificial Intelligence (AI) in Molecular Docking
Artificial Intelligence is the use of machines to replicate or mimic human thinking, performing various tasks. In molecular docking studies, AI can be utilized to analyze vast amounts of data, such as genomic, proteomic, and chemical information, to identify potential drug molecules and predict drug efficacy or toxicity.
The main use of AI in molecular docking studies is to analyze the structural confirmation of the molecules to be utilized. Experimental techniques such as X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM) are utilised to obtain the structural confirmation of the molecules; these methods are time-consuming. There have been strides in overcoming this, and the software responsible for this was AlphaFold. This software helps in enhancing the fragment assembly technique using deep learning (DL) methods by utilising a deep residual convolutional neural network (CNN). This allows for effective capture of intricate patterns within the protein data.
Applications of Molecular Docking
Molecular docking has found its use in various sectors. Some of them are mentioned below:
Lead optimization
Due to its ability to predict an optimized orientation of biomolecules, it can be used to predict different binding modes of the ligand in the binding site of the target molecule. Such information can be further used to develop potent, selective, and efficient analogs.
Hit Identifications
Since molecular makes use of the scoring function and search methods, this can be used to screen huge online databases to retrieve potent biomolecules.
Drug-DNA Interactions Studies
A lot of the drugs available in the market utilize nucleic acids and auxiliary processes as their main cellular target. This is seen in the cases of anticancer therapeutic agents. By being able to study the correlation between a drug’s molecular structure and its cytotoxicity, rational design and synthesis of new drugs can be done.
ADMET prediction
Molecular docking studies can be used to predict the Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties of small molecules. These results can then be used to weed out compounds with unfavorable properties early in drug discovery and development.

Limitations of Molecular Docking
Despite its usefulness and accuracy, molecular docking is still far from perfect. Some of the limitations include:
Ligand and target preparation
Even though molecular docking software can predict known protein-bound poses with good accuracy of about 1.5-2 Å, with reported success rates in the range of 70–80%, there are still chances of discrepancies. If the biomolecules to be used are not prepared properly, this can lead to improper results, which can be a hindrance in the drug discovery process.
Structural conformation
Even though AlphaFold can help in speeding up the process of defining the target structure, it cannot be considered as the final result since machine learning works based on predictions. This can result in inaccurate results.
Handling of flexible protein receptor
The Protein can change its conformation based on the ligand that is dolce, resulting in a single conformation for that particular ligand. However, certain ligands would require different conformations in the protein to be able to bind efficiently; thus, there is a need to keep the receptor flexible. Although it is known that protein flexibility accounts for higher affinity to be reached between a given medication and its target, docking studies typically ignore the continual mobility of proteins between distinct conformational states with identical energies. One significant factor influencing the effectiveness of the conformational search is the number of degrees of freedom incorporated.
Conclusion
Molecular docking is a computational method that is utilized in the medicinal sector to study the interactions between various biomolecules. It is widely used in drug discovery to produce novel drugs through dry lab experiments before taking them to the wet lab. This conveniently cuts costs and the time taken for producing a drug, while increasing the chances of a drug being able to clear the clinical trials. Various docking software is available in the market to perform docking studies. All these software work in a standard procedure of target and ligand selection, preparation, docking using scoring functions and search methods, and then evaluation of the results. Molecular docking has found its applications for lead optimization, ADMET studies, and so on. Despite its challenges, molecular docking can prove a great stride in the future for drug discovery and development.
References
- Agarwal, S., & Mehrotra, R. J. J. C. (2016). An overview of molecular docking. JSM chem, 4(2), 1024-1028.
- Han, R., Yoon, H., Kim, G., Lee, H., & Lee, Y. (2023). Revolutionizing medicinal chemistry: the application of artificial intelligence (AI) in early drug discovery. Pharmaceuticals, 16(9), 1259.
- Shamim, S., Munawar, R., Rashid, Y., Qadar, S. M. Z., Bushra, R., Begum, I., … & Quds, T. (2024). Molecular docking: an insight from drug discovery to drug repurposing approach.
- Agu, P. C., Afiukwa, C. A., Orji, O. U., Ezeh, E. M., Ofoke, I. H., Ogbu, C. O., … & Aja, P. M. (2023). Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Scientific reports, 13(1), 13398.
- Raval, K., & Ganatra, T. (2022). Basics, types, and applications of molecular docking: A review. IP International Journal of Comprehensive and Advanced Pharmacology, 7(1), 12-16.
- Morris, G. M., & Lim-Wilby, M. (2008). Molecular docking. In Molecular modeling of proteins (pp. 365-382). Totowa, NJ: Humana Press.
- https://parssilico.com/blogs/86-top-10-molecular-docking-softwares
- https://www.cgl.ucsf.edu/chimera/docs/UsersGuide/tutorials/pdbintro.html
- Akhter, M. (2016). Challenges in docking: mini review. JSM Chem, 4(1025), 2334-1831.
