Microbe Notes is an educational niche blog/website related to microbiology (bacteriology, virology, parasitology, mycology, immunology, molecular biology, biochemistry, etc.) useful for biology and microbiology courses (High School, B.Sc, M.Sc., M.Phil., and Ph.D.).
Choose Notes Categories
Latest Notes
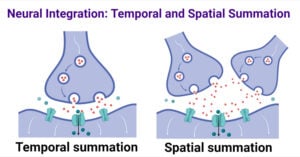
- Neural Integration: Temporal and Spatial Summation
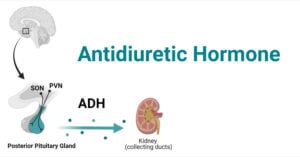
- Antidiuretic Hormone (ADH): Factors, Functions, Mechanism
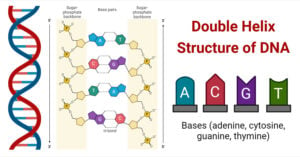
- Double Helix Structure of DNA with Diagram
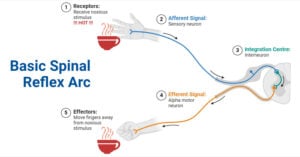
- Reflex Arc: Definition, Components, Functions
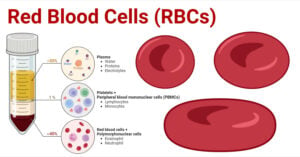
- Red Blood Cells (RBCs): Structure, Life Cycle, Functions
- Scientific Method: Definition, Steps, Examples, Uses
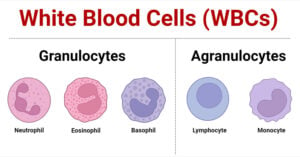
- White Blood Cells (WBCs): Types, Functions and Immunity
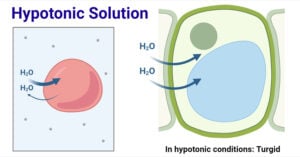
- Hypotonic Solution: Definition and Examples
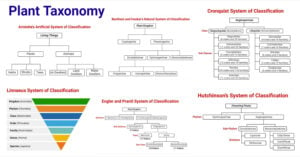
- Plant Taxonomy: Definition, Terms, Classifications
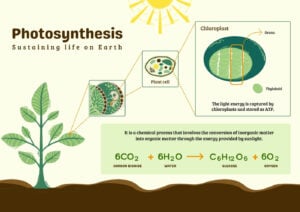
- Light Reaction and Dark Reaction of Photosynthesis
- Forest Biometrics: Definition, Tools, Methods, Uses
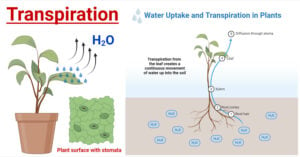
- Transpiration in Plants: Types, Mechanism, Factors, Uses
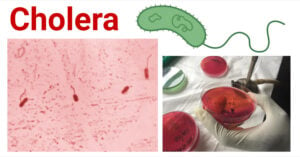
- Cholera: A-Level Biology Revision Notes
- Infectious Diseases: A-Level Biology Revision Notes
- Types of Plant Tissues: Meristematic and Permanent Tissue
- Simple Microscope: Principle, Parts, Uses, Examples, Diagram
- Sperm Cell: Anatomy, Structure, Functions, Diseases
- Forest Economics and Its Implications
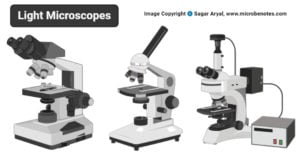
- Light Microscope: Principle, Types, Parts, Diagram
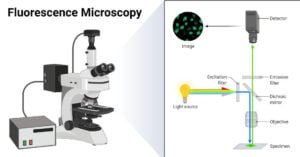
- Fluorescence Microscope: Principle, Parts, Uses, Examples